Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds
- PMID: 25200799
- PMCID: PMC4174253
- DOI: 10.1186/s12934-014-0126-z
Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds
Abstract
The production of aromatic amino acids using fermentation processes with recombinant microorganisms can be an advantageous approach to reach their global demands. In addition, a large array of compounds with alimentary and pharmaceutical applications can potentially be synthesized from intermediates of this metabolic pathway. However, contrary to other amino acids and primary metabolites, the artificial channelling of building blocks from central metabolism towards the aromatic amino acid pathway is complicated to achieve in an efficient manner. The length and complex regulation of this pathway have progressively called for the employment of more integral approaches, promoting the merge of complementary tools and techniques in order to surpass metabolic and regulatory bottlenecks. As a result, relevant insights on the subject have been obtained during the last years, especially with genetically modified strains of Escherichia coli. By combining metabolic engineering strategies with developments in synthetic biology, systems biology and bioprocess engineering, notable advances were achieved regarding the generation, characterization and optimization of E. coli strains for the overproduction of aromatic amino acids, some of their precursors and related compounds. In this paper we review and compare recent successful reports dealing with the modification of metabolic traits to attain these objectives.
Figures
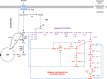
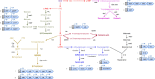
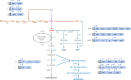
References
-
- Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martinez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muniz-Rascado L, Ong Q, Paley S, Schroder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. - DOI - PMC - PubMed
-
- Sprenger G. Aromatic Amino Acids. In: Wendisch VF, editor. Amin Acid Biosynth - Pathways, Regul Metab Eng. Berlin, Heidelberg: Springer; 2007. p. 418.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

