Involvement of p53 in the repair of DNA double strand breaks: multifaceted Roles of p53 in homologous recombination repair (HRR) and non-homologous end joining (NHEJ)
- PMID: 25201202
- PMCID: PMC4235614
- DOI: 10.1007/978-94-017-9211-0_17
Involvement of p53 in the repair of DNA double strand breaks: multifaceted Roles of p53 in homologous recombination repair (HRR) and non-homologous end joining (NHEJ)
Abstract
p53 is a tumor suppressor protein that prevents oncogenic transformation and maintains genomic stability by blocking proliferation of cells harboring unrepaired or misrepaired DNA. A wide range of genotoxic stresses such as DNA damaging anti-cancer drugs and ionizing radiation promote nuclear accumulation of p53 and trigger its ability to activate or repress a number of downstream target genes involved in various signaling pathways. This cascade leads to the activation of multiple cell cycle checkpoints and subsequent cell cycle arrest, allowing the cells to either repair the DNA or undergo apoptosis, depending on the intensity of DNA damage. In addition, p53 has many transcription-independent functions, including modulatory roles in DNA repair and recombination. This chapter will focus on the role of p53 in regulating or influencing the repair of DNA double-strand breaks that mainly includes homologous recombination repair (HRR) and non-homologous end joining (NHEJ). Through this discussion, we will try to establish that p53 acts as an important linchpin between upstream DNA damage signaling cues and downstream cellular events that include repair, recombination, and apoptosis.
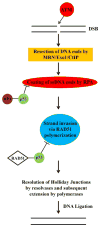
Figures

Similar articles
-
Proficiency in homologous recombination repair is prerequisite for activation of G2-checkpoint at low radiation doses.DNA Repair (Amst). 2021 May;101:103076. doi: 10.1016/j.dnarep.2021.103076. Epub 2021 Feb 20. DNA Repair (Amst). 2021. PMID: 33640756
-
Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations.Mutat Res Genet Toxicol Environ Mutagen. 2015 Nov;793:166-75. doi: 10.1016/j.mrgentox.2015.07.001. Epub 2015 Jul 4. Mutat Res Genet Toxicol Environ Mutagen. 2015. PMID: 26520387 Review.
-
Analysis of chromatid-break-repair detects a homologous recombination to non-homologous end-joining switch with increasing load of DNA double-strand breaks.Mutat Res Genet Toxicol Environ Mutagen. 2021 Jul;867:503372. doi: 10.1016/j.mrgentox.2021.503372. Epub 2021 Jun 12. Mutat Res Genet Toxicol Environ Mutagen. 2021. PMID: 34266628
-
Role for Artemis nuclease in the repair of radiation-induced DNA double strand breaks by alternative end joining.DNA Repair (Amst). 2015 Jul;31:29-40. doi: 10.1016/j.dnarep.2015.04.004. Epub 2015 Apr 25. DNA Repair (Amst). 2015. PMID: 25973742
-
Roles for 53BP1 in the repair of radiation-induced DNA double strand breaks.DNA Repair (Amst). 2020 Sep;93:102915. doi: 10.1016/j.dnarep.2020.102915. DNA Repair (Amst). 2020. PMID: 33087281 Review.
Cited by
-
Inactivation of RB1, CDKN2A, and TP53 have distinct effects on genomic stability at side-by-side comparison in karyotypically normal cells.Genes Chromosomes Cancer. 2023 Feb;62(2):93-100. doi: 10.1002/gcc.23096. Epub 2022 Sep 30. Genes Chromosomes Cancer. 2023. PMID: 36124964 Free PMC article.
-
Irradiation induces p53 loss of heterozygosity in breast cancer expressing mutant p53.Commun Biol. 2019 Nov 27;2:436. doi: 10.1038/s42003-019-0669-y. eCollection 2019. Commun Biol. 2019. PMID: 31799437 Free PMC article.
-
Physapruin A Enhances DNA Damage and Inhibits DNA Repair to Suppress Oral Cancer Cell Proliferation.Int J Mol Sci. 2022 Aug 9;23(16):8839. doi: 10.3390/ijms23168839. Int J Mol Sci. 2022. PMID: 36012104 Free PMC article.
-
Relationship between p53 status and the bioeffect of ionizing radiation.Oncol Lett. 2021 Sep;22(3):661. doi: 10.3892/ol.2021.12922. Epub 2021 Jul 14. Oncol Lett. 2021. PMID: 34386083 Free PMC article. Review.
-
MAPK15 upregulation promotes cell proliferation and prevents DNA damage in male germ cell tumors.Oncotarget. 2016 Apr 12;7(15):20981-98. doi: 10.18632/oncotarget.8044. Oncotarget. 2016. PMID: 26988910 Free PMC article.
References
-
- Albor A, Kaku S, Kulesz-Martin M. Wild-type and mutant forms of p53 activate human topoisomerase I: a possible mechanism for gain of function in mutants. Cancer research. 1998;58(10):2091–2094. - PubMed
-
- Amundson SA, Patterson A, Do KT, Fornace AJ., Jr A nucleotide excision repair master-switch: p53 regulated coordinate induction of global genomic repair genes. Cancer biology & therapy. 2002;1(2):145–149. - PubMed
-
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89(7):1175–1184. - PubMed
-
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science (New York, NY) 1998;281(5383):1674–1677. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

