Structural mechanisms in NLR inflammasome signaling
- PMID: 25201319
- PMCID: PMC4268015
- DOI: 10.1016/j.sbi.2014.08.011
Structural mechanisms in NLR inflammasome signaling
Abstract
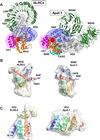
Members of the NOD-like receptor (NLR) family mediate the innate immune response to a wide range of pathogens, tissue damage and other cellular stresses. They achieve modulation of these signals by forming oligomeric signaling platforms, which in analogy to the apoptosome are predicted to adopt a defined oligomeric architecture and will here be referred to as NLR oligomers. Once formed, oligomers of the NLR proteins NLRP3 or NLRC4 'recruit' the adaptor protein ASC and the effector caspase-1, whereby NLRC4 can also directly interact with caspase-1. This results in large multi-protein assemblies, termed inflammasomes. Ultimately, the formation of these inflammasomes leads to the activation of caspase-1, which then processes the cytokines IL-1β and IL-18 triggering the immune response. Here we review new insights into NLR structure and implications on NLR oligomer formation as well as the nature of multi-protein inflammasomes. Of note, so dubbed 'canonical inflammasomes' can also be triggered by the NLR NLRP1b and the non-NLR protein AIM2, however the most detailed mechanistic information at hand pertains to NLRC4 while NLRP3 represents the quintessential inflammasome trigger. Thus these two NLRs are mainly used as examples in this article.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


References
-
- Lamkanfi M, Dixit VM, et al. Mechanisms and Functions of Inflammasomes. Cell. 2014;157:1013–1022. - PubMed
-
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. - PubMed
-
- Kersse K, Bertrand MJ, Lamkanfi M, Vandenabeele P. NOD-like receptors and the innate immune system: coping with danger, damage and death. Cytokine Growth Factor Rev. 2011;22:257–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

