Evolution of ibrutinib resistance in chronic lymphocytic leukemia (CLL)
- PMID: 25201956
- PMCID: PMC4183279
- DOI: 10.1073/pnas.1409362111
Evolution of ibrutinib resistance in chronic lymphocytic leukemia (CLL)
Abstract
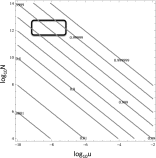
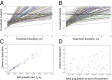
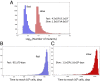
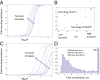
The Bruton tyrosine kinase inhibitor (BTKi) ibrutinib is a new targeted therapy for patients with chronic lymphocytic leukemia (CLL). Ibrutinib is given orally on a continuous schedule and induces durable remissions in the majority of CLL patients. However, a small proportion of patients initially responds to the BTKi and then develops resistance. Estimating the frequency, timing, and individual risk of developing resistance to ibrutinib, therefore, would be valuable for long-term management of patients. Computational evolutionary models, based on measured kinetic parameters of patients, allow us to approach these questions and to develop a roadmap for personalized prognosis and treatment management. Our kinetic models predict that BTKi-resistant mutants exist before initiation of ibrutinib therapy, although they only comprise a minority of the overall tumor burden. Furthermore, we can estimate the time required for resistant cells to grow to detectable levels. We predict that this can be highly variable, depending mostly on growth and death rates of the individual CLL cell clone. For a specific patient, this time can be predicted with a high degree of certainty. Our model can thus be used to predict for how long ibrutinib can suppress the disease in individual patients. Furthermore, the model can suggest whether prior debulking of the tumor with chemo-immunotherapy can prolong progression-free survival under ibrutinib. Finally, by applying the models to data that document progression during ibrutinib therapy, we estimated that resistant mutants might have a small (<2%) mean fitness advantage in the absence of treatment, compared with sensitive cells.
Keywords: drug resistance; evolutionary dynamics; mathematical models; personalized medicine; stochastic dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. - PubMed
-
- Pan Z, et al. Discovery of selective irreversible inhibitors for Bruton’s tyrosine kinase. ChemMedChem. 2007;2(1):58–61. - PubMed
-
- Burger JA, Buggy JJ. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) Leuk Lymphoma. 2013;54(11):2385–2391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

