A family of starch-active polysaccharide monooxygenases
- PMID: 25201969
- PMCID: PMC4183312
- DOI: 10.1073/pnas.1408090111
A family of starch-active polysaccharide monooxygenases
Abstract
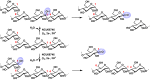
The recently discovered fungal and bacterial polysaccharide monooxygenases (PMOs) are capable of oxidatively cleaving chitin, cellulose, and hemicelluloses that contain β(1→4) linkages between glucose or substituted glucose units. They are also known collectively as lytic PMOs, or LPMOs, and individually as AA9 (formerly GH61), AA10 (formerly CBM33), and AA11 enzymes. PMOs share several conserved features, including a monocopper center coordinated by a bidentate N-terminal histidine residue and another histidine ligand. A bioinformatic analysis using these conserved features suggested several potential new PMO families in the fungus Neurospora crassa that are likely to be active on novel substrates. Herein, we report on NCU08746 that contains a C-terminal starch-binding domain and an N-terminal domain of previously unknown function. Biochemical studies showed that NCU08746 requires copper, oxygen, and a source of electrons to oxidize the C1 position of glycosidic bonds in starch substrates, but not in cellulose or chitin. Starch contains α(1→4) and α(1→6) linkages and exhibits higher order structures compared with chitin and cellulose. Cellobiose dehydrogenase, the biological redox partner of cellulose-active PMOs, can serve as the electron donor for NCU08746. NCU08746 contains one copper atom per protein molecule, which is likely coordinated by two histidine ligands as shown by X-ray absorption spectroscopy and sequence analysis. Results indicate that NCU08746 and homologs are starch-active PMOs, supporting the existence of a PMO superfamily with a much broader range of substrates. Starch-active PMOs provide an expanded perspective on studies of starch metabolism and may have potential in the food and starch-based biofuel industries.
Keywords: CBM20; copper enzymes; oxygen activation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hemsworth GR, Davies GJ, Walton PH. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol. 2013;23(5):660–668. - PubMed
-
- Berka RM, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol. 2011;29(10):922–927. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

