Medroxyprogesterone acetate differentially regulates interleukin (IL)-12 and IL-10 in a human ectocervical epithelial cell line in a glucocorticoid receptor (GR)-dependent manner
- PMID: 25202013
- PMCID: PMC4223317
- DOI: 10.1074/jbc.M114.587311
Medroxyprogesterone acetate differentially regulates interleukin (IL)-12 and IL-10 in a human ectocervical epithelial cell line in a glucocorticoid receptor (GR)-dependent manner
Abstract
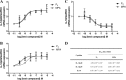
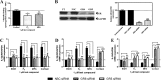
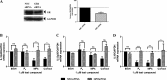
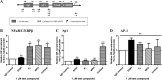
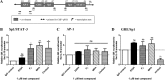
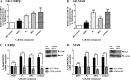
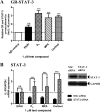
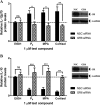
Medroxyprogesterone acetate (MPA), designed to mimic the actions of the endogenous hormone progesterone (P4), is extensively used by women as a contraceptive and in hormone replacement therapy. However, little is known about the steroid receptor-mediated molecular mechanisms of action of MPA in the female genital tract. In this study, we investigated the regulation of the pro-inflammatory cytokine, interleukin (IL)-12, and the anti-inflammatory cytokine IL-10, by MPA versus P4, in an in vitro cell culture model of the female ectocervical environment. This study shows that P4 and MPA significantly increase the expression of the IL-12p40 and IL-12p35 genes, whereas IL-10 gene expression is suppressed in a dose-dependent manner. Moreover, these effects were abrogated when reducing the glucocorticoid receptor (GR) levels with siRNA. Using a combination of chromatin immunoprecipitation (ChIP), siRNA, and re-ChIP assays, we show that recruitment of the P4- and MPA-bound GR to the IL-12p40 promoter requires CCAAT enhancer-binding protein (C/EBP)-β and nuclear factor κB (NFκB), although recruitment to the IL-10 promoter requires signal transducer and activator of transcription (STAT)-3. These results suggest that both P4 and MPA may modulate inflammation in the ectocervix via this genomic mechanism.
Keywords: Cervical Epithelium; Contraception; Cytokines; Gene Regulation; Glucocorticoid; Glucocorticoid Receptor; Inflammation; Medroxyprogesterone Acetate; Progesterone; Progestin.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Similar articles
-
The injectable-only contraceptive medroxyprogesterone acetate, unlike norethisterone acetate and progesterone, regulates inflammatory genes in endocervical cells via the glucocorticoid receptor.PLoS One. 2014 May 19;9(5):e96497. doi: 10.1371/journal.pone.0096497. eCollection 2014. PLoS One. 2014. PMID: 24840644 Free PMC article.
-
Differential regulation of endogenous pro-inflammatory cytokine genes by medroxyprogesterone acetate and norethisterone acetate in cell lines of the female genital tract.Contraception. 2011 Oct;84(4):423-35. doi: 10.1016/j.contraception.2011.06.006. Epub 2011 Aug 4. Contraception. 2011. PMID: 21920200
-
The progestin-only contraceptive medroxyprogesterone acetate, but not norethisterone acetate, enhances HIV-1 Vpr-mediated apoptosis in human CD4+ T cells through the glucocorticoid receptor.PLoS One. 2013 May 3;8(5):e62895. doi: 10.1371/journal.pone.0062895. Print 2013. PLoS One. 2013. PMID: 23658782 Free PMC article.
-
Differential glucocorticoid receptor-mediated effects on immunomodulatory gene expression by progestin contraceptives: implications for HIV-1 pathogenesis.Am J Reprod Immunol. 2014 Jun;71(6):505-12. doi: 10.1111/aji.12214. Epub 2014 Feb 18. Am J Reprod Immunol. 2014. PMID: 24547700 Review.
-
How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering.Mol Cell Endocrinol. 2013 Nov 5;380(1-2):41-54. doi: 10.1016/j.mce.2012.12.014. Epub 2012 Dec 23. Mol Cell Endocrinol. 2013. PMID: 23267834 Review.
Cited by
-
Stress, Sex, and Sugar: Glucocorticoids and Sex-Steroid Crosstalk in the Sex-Specific Misprogramming of Metabolism.J Endocr Soc. 2020 Jul 3;4(8):bvaa087. doi: 10.1210/jendso/bvaa087. eCollection 2020 Aug 1. J Endocr Soc. 2020. PMID: 32734132 Free PMC article. Review.
-
Medroxyprogesterone Acetate Impairs Amyloid Beta Degradation in a Matrix Metalloproteinase-9 Dependent Manner.Front Aging Neurosci. 2020 Apr 7;12:92. doi: 10.3389/fnagi.2020.00092. eCollection 2020. Front Aging Neurosci. 2020. PMID: 32317959 Free PMC article.
-
Glucocorticoids and Reproduction: Traffic Control on the Road to Reproduction.Trends Endocrinol Metab. 2017 Jun;28(6):399-415. doi: 10.1016/j.tem.2017.02.005. Epub 2017 Mar 6. Trends Endocrinol Metab. 2017. PMID: 28274682 Free PMC article. Review.
-
Differential off-target glucocorticoid activity of progestins used in endocrine therapy.Steroids. 2022 Jun;182:108998. doi: 10.1016/j.steroids.2022.108998. Epub 2022 Mar 7. Steroids. 2022. PMID: 35271867 Free PMC article.
-
Hormonal Contraception and HIV-1 Acquisition: Biological Mechanisms.Endocr Rev. 2018 Feb 1;39(1):36-78. doi: 10.1210/er.2017-00103. Endocr Rev. 2018. PMID: 29309550 Free PMC article. Review.
References
-
- Africander D., Verhoog N., Hapgood J. P. (2011) Molecular mechanisms of steroid receptor-mediated actions by synthetic progestins used in HRT and contraception. Steroids 76, 636–652 - PubMed
-
- Hapgood J. P., Koubovec D., Louw A., Africander D. (2004) Not all progestins are the same: implications for usage. Trends Pharmacol. Sci. 25, 554–557 - PubMed
-
- Speroff L., Darney P. D. (2010) in A Clinical Guide for Contraception (Seigafuse S., ed) 5th Ed., pp. 217–238, Lippincott Williams & Wilkins, Philadelphia
-
- Bailie R., Katzenellenbogen J., Hoffman M., Schierhout G., Truter H., Dent D., Gudgeon A., van Zyl J., Rosenberg L., Shapiro S. (1997) A case control study of breast cancer risk and exposure to injectable progestogen contraceptives. Methods and patterns of use among controls. S. Afr. Med. J. 87, 302–304 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

