DNA methylation analysis of SFRP2, GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in fecal DNA
- PMID: 25202404
- PMCID: PMC4156205
- DOI: 10.3892/ol.2014.2413
DNA methylation analysis of SFRP2, GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in fecal DNA
Abstract
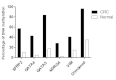
Aberrantly methylated genes are increasingly being established as biomarkers for the detection of colorectal cancer (CRC). In the present study, the methylation levels of the secreted frizzled-related protein gene 2 (SFRP2), GATA binding protein 4/5 (GATA4/5), N-Myc downstream-regulated gene 4 (NDRG4) and vimentin (VIM) promoters were evaluated for their use as markers in the noninvasive detection of CRC. Methylation-specific polymerase chain reaction was performed to analyze promoter CpG methylation of SFRP2, GATA4/5, NDRG4 and VIM in the fecal DNA of 56 patients with CRC and 40 individuals exhibiting normal colonoscopy results. Promoter methylation levels of SFRP2, GATA4/5, NDRG4 and VIM in CRC patients were 57.1% (32/56), 42.9% (24/56), 83.9% (47/56), 28.6% (16/56) and 41.1% (23/56), respectively. Furthermore, the specificity of the genes were 90.0% (4/40), 95.0% (2/40), 82.5% (7/40), 97.5% (4/40) and 85.0% (6/40), respectively. The overall sensitivity of detection for fecal DNA with at least one methylated gene was 96.4% (54/56) in CRC patients. By contrast, only 14 of the 40 normal individuals exhibited methylated DNA in the aforementioned promoter regions. Methylation of the SFRP2, GATA4/5, NDRG4 and VIM promoters in fecal DNA is associated with the presence of colorectal tumors. Therefore, the detection of aberrantly methylated DNA in fecal samples may present a promising, noninvasive screening method for CRC.
Keywords: colorectal cancer; fecal DNA; methylated DNA.
Figures

Similar articles
-
DNA methylation analysis of secreted frizzled-related protein 2 gene for the early detection of colorectal cancer in fecal DNA.Niger Med J. 2016 Jul-Aug;57(4):242-5. doi: 10.4103/0300-1652.188357. Niger Med J. 2016. PMID: 27630389 Free PMC article.
-
Is methylation analysis of SFRP2, TFPI2, NDRG4, and BMP3 promoters suitable for colorectal cancer screening in the Korean population?Intest Res. 2017 Oct;15(4):495-501. doi: 10.5217/ir.2017.15.4.495. Epub 2017 Oct 23. Intest Res. 2017. PMID: 29142517 Free PMC article.
-
Field Cancerization in Sporadic Colon Cancer.Gut Liver. 2016 Sep 15;10(5):773-80. doi: 10.5009/gnl15334. Gut Liver. 2016. PMID: 27114416 Free PMC article.
-
A systematic review and quantitative assessment of methylation biomarkers in fecal DNA and colorectal cancer and its precursor, colorectal adenoma.Mutat Res Rev Mutat Res. 2019 Jan-Mar;779:45-57. doi: 10.1016/j.mrrev.2019.01.003. Epub 2019 Jan 16. Mutat Res Rev Mutat Res. 2019. PMID: 31097151
-
Fecal DNA methylation markers for detecting stages of colorectal cancer and its precursors: a systematic review.Clin Epigenetics. 2020 Aug 10;12(1):122. doi: 10.1186/s13148-020-00904-7. Clin Epigenetics. 2020. PMID: 32778176 Free PMC article.
Cited by
-
Multigenerational epigenetic inheritance in humans: DNA methylation changes associated with maternal exposure to lead can be transmitted to the grandchildren.Sci Rep. 2015 Sep 29;5:14466. doi: 10.1038/srep14466. Sci Rep. 2015. PMID: 26417717 Free PMC article.
-
NDRG4 hypermethylation is a potential biomarker for diagnosis and prognosis of gastric cancer in Chinese population.Oncotarget. 2017 Jan 31;8(5):8105-8119. doi: 10.18632/oncotarget.14099. Oncotarget. 2017. PMID: 28042954 Free PMC article.
-
Identification of DNA methylation markers for early detection of CRC indicates a role for nervous system-related genes in CRC.Clin Epigenetics. 2021 Apr 15;13(1):80. doi: 10.1186/s13148-021-01067-9. Clin Epigenetics. 2021. PMID: 33858496 Free PMC article.
-
Association between SFRP promoter hypermethylation and different types of cancer: A systematic review and meta-analysis.Oncol Lett. 2019 Oct;18(4):3481-3492. doi: 10.3892/ol.2019.10709. Epub 2019 Aug 2. Oncol Lett. 2019. PMID: 31516566 Free PMC article.
-
Epigenetic and metabolic reprogramming in inflammatory bowel diseases: diagnostic and prognostic biomarkers in colorectal cancer.Cancer Cell Int. 2023 Nov 7;23(1):264. doi: 10.1186/s12935-023-03117-z. Cancer Cell Int. 2023. PMID: 37936149 Free PMC article. Review.
References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. - PubMed
-
- Jenkinson F, Steele RJ. Colorectal cancer screening-methodology. Surgeon. 2010;8:164–171. - PubMed
-
- Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. - PubMed
-
- Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. - PubMed
-
- Baek YH, Chang E, Kim YJ, et al. Stool methylation-specific polymerase chain reaction assay for the detection of colorectal neoplasia in Korean patients. Dis Colon Rectum. 2009;52:1452–1459. discussion 1459–1463. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
