Vulnerabilities of mutant SWI/SNF complexes in cancer
- PMID: 25203320
- PMCID: PMC4159614
- DOI: 10.1016/j.ccr.2014.07.018
Vulnerabilities of mutant SWI/SNF complexes in cancer
Abstract
Cancer genome sequencing efforts have revealed the novel theme that chromatin modifiers are frequently mutated across a wide spectrum of cancers. Mutations in genes encoding subunits of SWI/SNF (BAF) chromatin remodeling complexes are particularly prevalent, occurring in 20% of all human cancers. As these are typically loss-of-function mutations and not directly therapeutically targetable, central goals have been to elucidate mechanism and identify vulnerabilities created by these mutations. Here, we discuss emerging data that these mutations lead to the formation of aberrant residual SWI/SNF complexes that constitute a specific vulnerability and discuss the potential for exploiting these dependencies in SWI/SNF mutant cancers.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
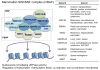
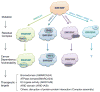
References
-
- Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene. 2002;21:5193–5203. - PubMed
-
- Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. - PubMed
-
- Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, Magnuson T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

