The landscape of kinase fusions in cancer
- PMID: 25204415
- PMCID: PMC4175590
- DOI: 10.1038/ncomms5846
The landscape of kinase fusions in cancer
Abstract
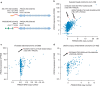
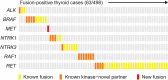
Human cancer genomes harbour a variety of alterations leading to the deregulation of key pathways in tumour cells. The genomic characterization of tumours has uncovered numerous genes recurrently mutated, deleted or amplified, but gene fusions have not been characterized as extensively. Here we develop heuristics for reliably detecting gene fusion events in RNA-seq data and apply them to nearly 7,000 samples from The Cancer Genome Atlas. We thereby are able to discover several novel and recurrent fusions involving kinases. These findings have immediate clinical implications and expand the therapeutic options for cancer patients, as approved or exploratory drugs exist for many of these kinases.
Conflict of interest statement
All authors of this manuscript are employees and shareholders of Blueprint Medicines.
Figures



References
-
- Shaw A. T. et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. New Engl. J. Med. 368, 2385–2394 (2013). - PubMed
-
- Soda M. et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007). - PubMed
-
- Lin E. et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol. Cancer Res. 7, 1466–1476 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

