Single-molecule correlated chemical probing of RNA
- PMID: 25205807
- PMCID: PMC4183288
- DOI: 10.1073/pnas.1407306111
Single-molecule correlated chemical probing of RNA
Abstract
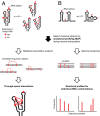
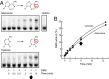
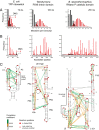
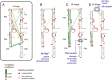
Complex higher-order RNA structures play critical roles in all facets of gene expression; however, the through-space interaction networks that define tertiary structures and govern sampling of multiple conformations are poorly understood. Here we describe single-molecule RNA structure analysis in which multiple sites of chemical modification are identified in single RNA strands by massively parallel sequencing and then analyzed for correlated and clustered interactions. The strategy thus identifies RNA interaction groups by mutational profiling (RING-MaP) and makes possible two expansive applications. First, we identify through-space interactions, create 3D models for RNAs spanning 80-265 nucleotides, and characterize broad classes of intramolecular interactions that stabilize RNA. Second, we distinguish distinct conformations in solution ensembles and reveal previously undetected hidden states and large-scale structural reconfigurations that occur in unfolded RNAs relative to native states. RING-MaP single-molecule nucleic acid structure interrogation enables concise and facile analysis of the global architectures and multiple conformations that govern function in RNA.
Keywords: dimethyl sulfate; motif discovery; spectral clustering; structure refinement.
Conflict of interest statement
Conflict of interest statement: The authors have applied for a provisional patent on elements of this work.
Figures


References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

