Determinants for tRNA-dependent pretransfer editing in the synthetic site of isoleucyl-tRNA synthetase
- PMID: 25207837
- PMCID: PMC4188249
- DOI: 10.1021/bi5007699
Determinants for tRNA-dependent pretransfer editing in the synthetic site of isoleucyl-tRNA synthetase
Abstract
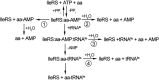
The accurate expression of genetic information relies on the fidelity of amino acid-tRNA coupling by aminoacyl-tRNA synthetases (aaRS). When the specificity against structurally similar noncognate amino acids in the synthetic reaction does not support a threshold fidelity level for translation, the aaRS employ intrinsic hydrolytic editing to correct errors in aminoacylation. Escherichia coli isoleucyl-tRNA synthetase (EcIleRS) is a class I aaRS that is notable for its use of tRNA-dependent pretransfer editing to hydrolyze noncognate valyl-adenylate prior to aminoacyl-tRNA formation. On the basis of the finding that IleRS possessing an inactivated post-transfer editing domain is still capable of robust tRNA-dependent editing, we have recently proposed that the pretransfer editing activity resides within the synthetic site. Here we apply an improved methodology that allows quantitation of the AMP fraction that arises particularly from tRNA-dependent aa-AMP hydrolysis. By this approach, we demonstrate that tRNA-dependent pretransfer editing accounts for nearly one-third of the total proofreading by EcIleRS and that a highly conserved tyrosine within the synthetic site modulates both editing and aminoacylation. Therefore, synthesis of aminoacyl-tRNA and hydrolysis of aminoacyl-adenylates employ overlapping amino acid determinants. We suggest that this overlap hindered the evolution of synthetic site-based pretransfer editing as the predominant proofreading pathway, because that activity is difficult to accommodate in the context of efficient aminoacyl-tRNA synthesis. Instead, the acquisition of a spatially separate domain dedicated to post-transfer editing alone allowed for the development of a powerful deacylation machinery that effectively competes with dissociation of misacylated tRNAs.
Figures

References
-
- First E. A. (2005) Catalysis of the tRNA aminoacylation reaction. In The aminoacyl-tRNA synthetases (Ibba M., Francklyn C. S., and Cusack S., Eds.) pp 328–347, Landes Bioscience/Eurekah.com, Georgetown, TX.
-
- Perona J. J.; Hadd A. (2012) Structural diversity and protein engineering of the aminoacyl-tRNA synthetases. Biochemistry 51, 8705–8729. - PubMed
-
- Eriani G.; Delarue M.; Poch O.; Gangloff J.; Moras D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347, 203–206. - PubMed
-
- Zhang C. M.; Perona J. J.; Ryu K.; Francklyn C.; Hou Y. M. (2006) Distinct kinetic mechanisms of the two classes of aminoacyl-tRNA synthetases. J. Mol. Biol. 361, 300–311. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

