Engineering amyloid-like assemblies from unstructured peptides via site-specific lipid conjugation
- PMID: 25207975
- PMCID: PMC4160191
- DOI: 10.1371/journal.pone.0105641
Engineering amyloid-like assemblies from unstructured peptides via site-specific lipid conjugation
Abstract
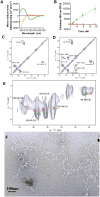
Aggregation of amyloid beta (Aβ) into oligomers and fibrils is believed to play an important role in the development of Alzheimer's disease (AD). To gain further insight into the principles of aggregation, we have investigated the induction of β-sheet secondary conformation from disordered native peptide sequences through lipidation, in 1-2% hexafluoroisopropanol (HFIP) in phosphate buffered saline (PBS). Several parameters, such as type and number of lipid chains, peptide sequence, peptide length and net charge, were explored keeping the ratio peptide/HFIP constant. The resulting lipoconjugates were characterized by several physico-chemical techniques: Circular Dichroism (CD), Attenuated Total Reflection InfraRed (ATR-IR), Thioflavin T (ThT) fluorescence, Dynamic Light Scattering (DLS), solid-state Nuclear Magnetic Resonance (ssNMR) spectroscopy and Electron Microscopy (EM). Our data demonstrate the generation of β-sheet aggregates from numerous unstructured peptides under physiological pH, independent of the amino acid sequence. The amphiphilicity pattern and hydrophobicity of the scaffold were found to be key factors for their assembly into amyloid-like structures.
Conflict of interest statement
Figures

Similar articles
-
Aggregation pathways of the amyloid β(1-42) peptide depend on its colloidal stability and ordered β-sheet stacking.Langmuir. 2012 Sep 4;28(35):12711-21. doi: 10.1021/la3021436. Epub 2012 Aug 22. Langmuir. 2012. PMID: 22870885 Free PMC article.
-
N-terminal lipid conjugation of amyloid β(1-40) leads to the formation of highly ordered N-terminally extended fibrils.Phys Chem Chem Phys. 2017 Jan 18;19(3):1839-1846. doi: 10.1039/c6cp05982a. Phys Chem Chem Phys. 2017. PMID: 28000812
-
The Japanese mutant Aβ (ΔE22-Aβ(1-39)) forms fibrils instantaneously, with low-thioflavin T fluorescence: seeding of wild-type Aβ(1-40) into atypical fibrils by ΔE22-Aβ(1-39).Biochemistry. 2011 Mar 29;50(12):2026-39. doi: 10.1021/bi1016217. Epub 2011 Feb 24. Biochemistry. 2011. PMID: 21291268 Free PMC article.
-
Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases.Acc Chem Res. 2018 Mar 20;51(3):706-718. doi: 10.1021/acs.accounts.7b00554. Epub 2018 Mar 6. Acc Chem Res. 2018. PMID: 29508987 Free PMC article. Review.
-
ATR-FTIR: a "rejuvenated" tool to investigate amyloid proteins.Biochim Biophys Acta. 2013 Oct;1828(10):2328-38. doi: 10.1016/j.bbamem.2013.04.012. Epub 2013 Jun 5. Biochim Biophys Acta. 2013. PMID: 23746423 Review.
Cited by
-
Unnatural Amino Acid: 4-Aminopyrazolonyl Amino Acid Comprising Tri-Peptides Forms Organogel With Co-Solvent (EtOAc:Hexane).Front Chem. 2022 May 5;10:821971. doi: 10.3389/fchem.2022.821971. eCollection 2022. Front Chem. 2022. PMID: 35601543 Free PMC article.
References
-
- Forns P, Lauer-Fields JL, Gao S, Fields GB (2000) Induction of protein-like molecular architecture by monoalkyl hydrocarbon chains. Biopolymers 54: 531–546. - PubMed
-
- Krug M, Folkers G, Haas B, Hess G, Wiesmuller KH, et al. (1989) Molecular dynamics of the alpha-helical epitope of a novel synthetic lipopeptide foot-and-mouth disease virus vaccine. Biopolymers 28: 499–512. - PubMed
-
- Yu Y-C, Tirrell M, Fields GB (1998) Minimal Lipidation Stabilizes Protein-Like Molecular Architecture. J Am Chem Soc 120: 9979–9987.
-
- Paramonov SE, Jun HW, Hartgerink JD (2006) Self-assembly of peptide-amphiphile nanofibers: the roles of hydrogen bonding and amphiphilic packing. J Am Chem Soc 128: 7291–7298. - PubMed
-
- Cavalli S, Handgraaf JW, Tellers EE, Popescu DC, Overhand M, et al. (2006) Two-dimensional ordered beta-sheet lipopeptide monolayers. J Am Chem Soc 128: 13959–13966. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

