A comparative metagenome survey of the fecal microbiota of a breast- and a plant-fed Asian elephant reveals an unexpectedly high diversity of glycoside hydrolase family enzymes
- PMID: 25208077
- PMCID: PMC4160196
- DOI: 10.1371/journal.pone.0106707
A comparative metagenome survey of the fecal microbiota of a breast- and a plant-fed Asian elephant reveals an unexpectedly high diversity of glycoside hydrolase family enzymes
Abstract
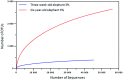
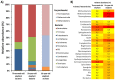
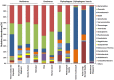
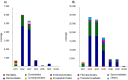
A phylogenetic and metagenomic study of elephant feces samples (derived from a three-weeks-old and a six-years-old Asian elephant) was conducted in order to describe the microbiota inhabiting this large land-living animal. The microbial diversity was examined via 16S rRNA gene analysis. We generated more than 44,000 GS-FLX+454 reads for each animal. For the baby elephant, 380 operational taxonomic units (OTUs) were identified at 97% sequence identity level; in the six-years-old animal, close to 3,000 OTUs were identified, suggesting high microbial diversity in the older animal. In both animals most OTUs belonged to Bacteroidetes and Firmicutes. Additionally, for the baby elephant a high number of Proteobacteria was detected. A metagenomic sequencing approach using Illumina technology resulted in the generation of 1.1 Gbp assembled DNA in contigs with a maximum size of 0.6 Mbp. A KEGG pathway analysis suggested high metabolic diversity regarding the use of polymers and aromatic and non-aromatic compounds. In line with the high phylogenetic diversity, a surprising and not previously described biodiversity of glycoside hydrolase (GH) genes was found. Enzymes of 84 GH families were detected. Polysaccharide utilization loci (PULs), which are found in Bacteroidetes, were highly abundant in the dataset; some of these comprised cellulase genes. Furthermore the highest coverage for GH5 and GH9 family enzymes was detected for Bacteroidetes, suggesting that bacteria of this phylum are mainly responsible for the degradation of cellulose in the Asian elephant. Altogether, this study delivers insight into the biomass conversion by one of the largest plant-fed and land-living animals.
Conflict of interest statement
Figures

Similar articles
-
Metagenomic insights into the diversity of carbohydrate-degrading enzymes in the yak fecal microbial community.BMC Microbiol. 2020 Oct 10;20(1):302. doi: 10.1186/s12866-020-01993-3. BMC Microbiol. 2020. PMID: 33036549 Free PMC article.
-
Comparative and functional analyses of fecal microbiome in Asian elephants.Antonie Van Leeuwenhoek. 2022 Sep;115(9):1187-1202. doi: 10.1007/s10482-022-01757-1. Epub 2022 Jul 28. Antonie Van Leeuwenhoek. 2022. PMID: 35902439
-
Metagenomic analysis of the Rhinopithecus bieti fecal microbiome reveals a broad diversity of bacterial and glycoside hydrolase profiles related to lignocellulose degradation.BMC Genomics. 2015 Mar 12;16(1):174. doi: 10.1186/s12864-015-1378-7. BMC Genomics. 2015. PMID: 25887697 Free PMC article.
-
Metagenomic Analysis of the Fecal Microbiomes of Wild Asian Elephants Reveals Microflora and Enzymes that Mainly Digest Hemicellulose.J Microbiol Biotechnol. 2019 Aug 28;29(8):1255-1265. doi: 10.4014/jmb.1904.04033. J Microbiol Biotechnol. 2019. PMID: 31337187
-
Understanding the alteration in rumen microbiome and CAZymes profile with diet and host through comparative metagenomic approach.Arch Microbiol. 2019 Dec;201(10):1385-1397. doi: 10.1007/s00203-019-01706-z. Epub 2019 Jul 23. Arch Microbiol. 2019. PMID: 31338542
Cited by
-
A comprehensive survey of integron-associated genes present in metagenomes.BMC Genomics. 2020 Jul 20;21(1):495. doi: 10.1186/s12864-020-06830-5. BMC Genomics. 2020. PMID: 32689930 Free PMC article.
-
Assessing Metagenomic Signals Recovered from Lyuba, a 42,000-Year-Old Permafrost-Preserved Woolly Mammoth Calf.Genes (Basel). 2018 Aug 31;9(9):436. doi: 10.3390/genes9090436. Genes (Basel). 2018. PMID: 30200350 Free PMC article.
-
Analysis of infant microbiota composition and the relationship with breast milk components in the Asian elephant (Elephas maximus) at the zoo.J Vet Med Sci. 2020 Jul 31;82(7):983-989. doi: 10.1292/jvms.20-0190. Epub 2020 Apr 29. J Vet Med Sci. 2020. PMID: 32350162 Free PMC article.
-
Metagenomic insights into the diversity of carbohydrate-degrading enzymes in the yak fecal microbial community.BMC Microbiol. 2020 Oct 10;20(1):302. doi: 10.1186/s12866-020-01993-3. BMC Microbiol. 2020. PMID: 33036549 Free PMC article.
-
Biochemical Characteristics of Microbial Enzymes and Their Significance from Industrial Perspectives.Mol Biotechnol. 2019 Aug;61(8):579-601. doi: 10.1007/s12033-019-00187-1. Mol Biotechnol. 2019. PMID: 31168761 Review.
References
-
- Björnhag G (1994) The Digestive System in Mammals: Food, Form and Function: Cambridge University Press: 287–309.
-
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA (2008) Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature Reviews Microbiology 6: 121–131. - PubMed
-
- Bayané A, Guiot S (2011) Animal digestive strategies versus anaerobic digestion bioprocesses for biogas production from lignocellulosic biomass. Reviews in Environmental Science and Bio/Technology 10: 43–62.
-
- Clauss M, Frey R, Kiefer B, Lechner-Doll M, Loehlein W, et al. (2003) The maximum attainable body size of herbivorous mammals: morphophysiological constraints on foregut, and adaptations of hindgut fermenters. Oecologia 136: 14–27. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

