Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer
- PMID: 25208879
- PMCID: PMC4362882
- DOI: 10.1158/1078-0432.CCR-14-0432
Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer
Abstract
Purpose: Genomic profiling studies suggest that triple-negative breast cancer (TNBC) is a heterogeneous disease. In this study, we sought to define TNBC subtypes and identify subtype-specific markers and targets.
Experimental design: RNA and DNA profiling analyses were conducted on 198 TNBC tumors [estrogen receptor (ER) negativity defined as Allred scale value ≤ 2] with >50% cellularity (discovery set: n = 84; validation set: n = 114) collected at Baylor College of Medicine (Houston, TX). An external dataset of seven publically accessible TNBC studies was used to confirm results. DNA copy number, disease-free survival (DFS), and disease-specific survival (DSS) were analyzed independently using these datasets.
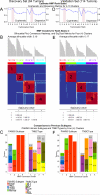
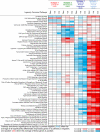
Results: We identified and confirmed four distinct TNBC subtypes: (i) luminal androgen receptor (AR; LAR), (ii) mesenchymal (MES), (iii) basal-like immunosuppressed (BLIS), and (iv) basal-like immune-activated (BLIA). Of these, prognosis is worst for BLIS tumors and best for BLIA tumors for both DFS (log-rank test: P = 0.042 and 0.041, respectively) and DSS (log-rank test: P = 0.039 and 0.029, respectively). DNA copy number analysis produced two major groups (LAR and MES/BLIS/BLIA) and suggested that gene amplification drives gene expression in some cases [FGFR2 (BLIS)]. Putative subtype-specific targets were identified: (i) LAR: androgen receptor and the cell surface mucin MUC1, (ii) MES: growth factor receptors [platelet-derived growth factor (PDGF) receptor A; c-Kit], (iii) BLIS: an immunosuppressing molecule (VTCN1), and (iv) BLIA: Stat signal transduction molecules and cytokines.
Conclusion: There are four stable TNBC subtypes characterized by the expression of distinct molecular profiles that have distinct prognoses. These studies identify novel subtype-specific targets that can be targeted in the future for the effective treatment of TNBCs.
©2014 American Association for Cancer Research.
Figures


Comment in
-
Translating the molecular message of triple-negative breast cancer into targeted therapy.Clin Cancer Res. 2015 Apr 1;21(7):1511-3. doi: 10.1158/1078-0432.CCR-14-2532. Epub 2014 Oct 28. Clin Cancer Res. 2015. PMID: 25351747
References
-
- Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005 Oct 10;23(29):7350–60. - PubMed
-
- Morris GJ, Naidu S, Topham AK, Guiles F, Xu Y, McCue P, et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: a single-institution compilation compared with the National Cancer Institute's Surveillance, Epidemiology, and End Results database. Cancer. 2007 Aug 15;110(4):876–84. - PubMed
-
- Shastry M, Yardley DA. Updates in the treatment of basal/triple-negative breast cancer. Curr Opin Obstet Gynecol. 2013 Feb;25(1):40–8. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

