Inactivation of agmatinase expressed in vegetative cells alters arginine catabolism and prevents diazotrophic growth in the heterocyst-forming cyanobacterium Anabaena
- PMID: 25209059
- PMCID: PMC4234267
- DOI: 10.1002/mbo3.207
Inactivation of agmatinase expressed in vegetative cells alters arginine catabolism and prevents diazotrophic growth in the heterocyst-forming cyanobacterium Anabaena
Abstract
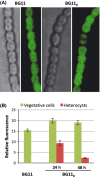
Arginine decarboxylase produces agmatine, and arginase and agmatinase are ureohydrolases that catalyze the production of ornithine and putrescine from arginine and agmatine, respectively, releasing urea. In the genome of the filamentous, heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120, ORF alr2310 putatively encodes an ureohydrolase. Cells of Anabaena supplemented with [(14) C]arginine took up and catabolized this amino acid generating a set of labeled amino acids that included ornithine, proline, and glutamate. In an alr2310 deletion mutant, an agmatine spot appeared and labeled glutamate increased with respect to the wild type, suggesting that Alr2310 is an agmatinase rather than an arginase. As determined in cell-free extracts, agmatinase activity could be detected in the wild type but not in the mutant. Thus, alr2310 is the Anabaena speB gene encoding agmatinase. The ∆alr2310 mutant accumulated large amounts of cyanophycin granule polypeptide, lacked nitrogenase activity, and did not grow diazotrophically. Growth tests in solid media showed that agmatine is inhibitory for Anabaena, especially under diazotrophic conditions, suggesting that growth of the mutant is inhibited by non-metabolized agmatine. Measurements of incorporation of radioactivity from [(14) C]leucine into macromolecules showed, however, a limited inhibition of protein synthesis in the ∆alr2310 mutant. Analysis of an Anabaena strain producing an Alr2310-GFP (green fluorescent protein) fusion showed expression in vegetative cells but much less in heterocysts, implying compartmentalization of the arginine decarboxylation pathway in the diazotrophic filaments of this heterocyst-forming cyanobacterium.
Keywords: Agmatinase; Anabaena; arginine catabolism; nitrogenase.
© 2014 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures






Similar articles
-
Compartmentalized cyanophycin metabolism in the diazotrophic filaments of a heterocyst-forming cyanobacterium.Proc Natl Acad Sci U S A. 2014 Mar 11;111(10):3823-8. doi: 10.1073/pnas.1318564111. Epub 2014 Feb 18. Proc Natl Acad Sci U S A. 2014. PMID: 24550502 Free PMC article.
-
Homospermidine biosynthesis in the cyanobacterium Anabaena requires a deoxyhypusine synthase homologue and is essential for normal diazotrophic growth.Mol Microbiol. 2018 Sep;109(6):763-780. doi: 10.1111/mmi.14006. Epub 2018 Oct 4. Mol Microbiol. 2018. PMID: 29923645
-
Catabolic function of compartmentalized alanine dehydrogenase in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120.J Bacteriol. 2010 Oct;192(19):5165-72. doi: 10.1128/JB.00603-10. Epub 2010 Jul 30. J Bacteriol. 2010. PMID: 20675483 Free PMC article.
-
Heterocyst Septa Contain Large Nanopores That Are Influenced by the Fra Proteins in the Filamentous Cyanobacterium Anabaena sp. Strain PCC 7120.J Bacteriol. 2021 Jun 8;203(13):e0008121. doi: 10.1128/JB.00081-21. Epub 2021 Jun 8. J Bacteriol. 2021. PMID: 33846119 Free PMC article.
-
Vertebrate agmatinases: what role do they play in agmatine catabolism?Ann N Y Acad Sci. 2003 Dec;1009:30-3. doi: 10.1196/annals.1304.003. Ann N Y Acad Sci. 2003. PMID: 15028567 Review.
Cited by
-
Discovery of a Ni2+-dependent guanidine hydrolase in bacteria.Nature. 2022 Mar;603(7901):515-521. doi: 10.1038/s41586-022-04490-x. Epub 2022 Mar 9. Nature. 2022. PMID: 35264792
-
Extracellular Streptomyces lividans vesicles: composition, biogenesis and antimicrobial activity.Microb Biotechnol. 2015 Jul;8(4):644-58. doi: 10.1111/1751-7915.12274. Epub 2015 Apr 7. Microb Biotechnol. 2015. PMID: 25851532 Free PMC article.
-
Large-scale phylogenomics of aquatic bacteria reveal molecular mechanisms for adaptation to salinity.Sci Adv. 2023 May 26;9(21):eadg2059. doi: 10.1126/sciadv.adg2059. Epub 2023 May 26. Sci Adv. 2023. PMID: 37235649 Free PMC article.
-
ZipN is an essential FtsZ membrane tether and contributes to the septal localization of SepJ in the filamentous cyanobacterium Anabaena.Sci Rep. 2019 Feb 26;9(1):2744. doi: 10.1038/s41598-019-39336-6. Sci Rep. 2019. PMID: 30808920 Free PMC article.
-
The virulence factor urease and its unexplored role in the metabolism of Cryptococcus neoformans.FEMS Yeast Res. 2020 Jun 1;20(4):foaa031. doi: 10.1093/femsyr/foaa031. FEMS Yeast Res. 2020. PMID: 32490521 Free PMC article.
References
-
- Berg H, Ziegler K, Piotukh K, Baier K, Lockau W. Volkmer-Engert R. Biosynthesis of the cyanobacterial reserve polymer multi-L-arginyl-poly-L-aspartic acid (cyanophycin): mechanism of the cyanophycin synthetase reaction studied with synthetic primers. Eur. J. Biochem. 2000;267:5561–5570. - PubMed
-
- Boyde TRC. Rahmatullah M. Optimization of conditions for the colorimetric determination of citrulline, using diacetyl monoxime. Anal. Biochem. 1980;107:424–431. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

