The splicing factor PQBP1 regulates mesodermal and neural development through FGF signaling
- PMID: 25209246
- PMCID: PMC4197583
- DOI: 10.1242/dev.106658
The splicing factor PQBP1 regulates mesodermal and neural development through FGF signaling
Abstract
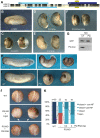
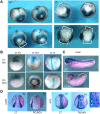
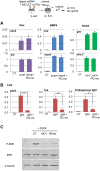
Alternative splicing of pre-mRNAs is an important means of regulating developmental processes, yet the molecular mechanisms governing alternative splicing in embryonic contexts are just beginning to emerge. Polyglutamine-binding protein 1 (PQBP1) is an RNA-splicing factor that, when mutated, in humans causes Renpenning syndrome, an X-linked intellectual disability disease characterized by severe cognitive impairment, but also by physical defects that suggest PQBP1 has broader functions in embryonic development. Here, we reveal essential roles for PQBP1 and a binding partner, WBP11, in early development of Xenopus embryos. Both genes are expressed in the nascent mesoderm and neurectoderm, and morpholino knockdown of either causes defects in differentiation and morphogenesis of the mesoderm and neural plate. At the molecular level, knockdown of PQBP1 in Xenopus animal cap explants inhibits target gene induction by FGF but not by BMP, Nodal or Wnt ligands, and knockdown of either PQBP1 or WBP11 in embryos inhibits expression of fgf4 and FGF4-responsive cdx4 genes. Furthermore, PQBP1 knockdown changes the alternative splicing of FGF receptor-2 (FGFR2) transcripts, altering the incorporation of cassette exons that generate receptor variants (FGFR2 IIIb or IIIc) with different ligand specificities. Our findings may inform studies into the mechanisms underlying Renpenning syndrome.
Keywords: Alternative splicing; FGF; FGF receptor; Mesoderm; Neural; PQBP1; Renpenning syndrome; WBP11; Xenopus.
© 2014. Published by The Company of Biologists Ltd.
Figures




Similar articles
-
fus/TLS orchestrates splicing of developmental regulators during gastrulation.Genes Dev. 2012 Jun 15;26(12):1351-63. doi: 10.1101/gad.187278.112. Genes Dev. 2012. PMID: 22713872 Free PMC article.
-
Identification and characterization of Xenopus kctd15, an ectodermal gene repressed by the FGF pathway.Int J Dev Biol. 2012;56(5):393-402. doi: 10.1387/ijdb.113333ct. Int J Dev Biol. 2012. PMID: 22811273
-
The nodal target gene Xmenf is a component of an FGF-independent pathway of ventral mesoderm induction in Xenopus.Mech Dev. 2002 Oct;118(1-2):45-56. doi: 10.1016/s0925-4773(02)00186-7. Mech Dev. 2002. PMID: 12351169
-
The role of PQBP1 in neural development and function.Biochem Soc Trans. 2023 Feb 27;51(1):363-372. doi: 10.1042/BST20220920. Biochem Soc Trans. 2023. PMID: 36815699 Review.
-
Role of PQBP1 in Pathogen Recognition-Impact on Innate Immunity.Viruses. 2024 Aug 21;16(8):1340. doi: 10.3390/v16081340. Viruses. 2024. PMID: 39205314 Free PMC article. Review.
Cited by
-
Expanding the genetic toolkit in Xenopus: Approaches and opportunities for human disease modeling.Dev Biol. 2017 Jun 15;426(2):325-335. doi: 10.1016/j.ydbio.2016.04.009. Epub 2016 Apr 22. Dev Biol. 2017. PMID: 27109192 Free PMC article. Review.
-
The emerging significance of splicing in vertebrate development.Development. 2022 Oct 1;149(19):dev200373. doi: 10.1242/dev.200373. Epub 2022 Sep 30. Development. 2022. PMID: 36178052 Free PMC article. Review.
-
Recognition of HIV-1 capsid by PQBP1 licenses an innate immune sensing of nascent HIV-1 DNA.Mol Cell. 2022 Aug 4;82(15):2871-2884.e6. doi: 10.1016/j.molcel.2022.06.010. Epub 2022 Jul 8. Mol Cell. 2022. PMID: 35809572 Free PMC article.
-
Prostate cancer resistance leads to a global deregulation of translation factors and unconventional translation.NAR Cancer. 2022 Nov 4;4(4):zcac034. doi: 10.1093/narcan/zcac034. eCollection 2022 Dec. NAR Cancer. 2022. PMID: 36348939 Free PMC article.
-
The miR-29c-KIAA1199 axis regulates gastric cancer migration by binding with WBP11 and PTP4A3.Oncogene. 2019 Apr;38(17):3134-3150. doi: 10.1038/s41388-018-0642-0. Epub 2019 Jan 9. Oncogene. 2019. PMID: 30626935
References
-
- Amaya, E., Stein, P. A., Musci, T. J. and Kirschner, M. W. (1993). FGF signalling in the early specification of mesoderm in Xenopus. Development 118, 477-487. - PubMed
-
- Blunt, A. G., Lawshé, A., Cunningham, M. L., Seto, M. L., Ornitz, D. M. and MacArthur, C. A. (1997). Overlapping expression and redundant activation of mesenchymal fibroblast growth factor (FGF) receptors by alternatively spliced FGF-8 ligands. J. Biol. Chem. 272, 3733-3738 10.1074/jbc.272.6.3733 - DOI - PubMed
-
- Busch, A., Engemann, S., Lurz, R., Okazawa, H., Lehrach, H. and Wanker, E. E. (2003). Mutant huntingtin promotes the fibrillogenesis of wild-type huntingtin: a potential mechanism for loss of huntingtin function in Huntington's disease. J. Biol. Chem. 278, 41452-41461 10.1074/jbc.M303354200 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

