Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells
- PMID: 25209314
- PMCID: PMC4217687
- DOI: 10.1093/cvr/cvu205
Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells
Abstract
Aims: Familial hypertrophic cardiomyopathy (HCM) is one the most common heart disorders, with gene mutations in the cardiac sarcomere. Studying HCM with patient-specific induced pluripotent stem-cell (iPSC)-derived cardiomyocytes (CMs) would benefit the understanding of HCM mechanism, as well as the development of personalized therapeutic strategies.
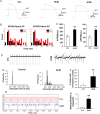
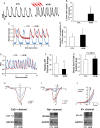
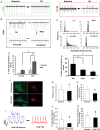
Methods and results: To investigate the molecular mechanism underlying the abnormal CM functions in HCM, we derived iPSCs from an HCM patient with a single missense mutation (Arginine442Glycine) in the MYH7 gene. CMs were next enriched from HCM and healthy iPSCs, followed with whole transcriptome sequencing and pathway enrichment analysis. A widespread increase of genes responsible for 'Cell Proliferation' was observed in HCM iPSC-CMs when compared with control iPSC-CMs. Additionally, HCM iPSC-CMs exhibited disorganized sarcomeres and electrophysiological irregularities. Furthermore, disease phenotypes of HCM iPSC-CMs were attenuated with pharmaceutical treatments.
Conclusion: Overall, this study explored the possible patient-specific and mutation-specific disease mechanism of HCM, and demonstrates the potential of using HCM iPSC-CMs for future development of therapeutic strategies. Additionally, the whole methodology established in this study could be utilized to study mechanisms of other human-inherited heart diseases.
Keywords: Cardiomyocyte; Heart; Hypertrophic cardiomyopathy; Induced pluripotent stem cells.
© The Author 2014. Published by Oxford University Press on behalf of the European Society of Cardiology.
Figures



Comment in
-
Reconciling computer models and stem cell models of human cardiac repolarization: reply.Cardiovasc Res. 2015 Apr 1;106(1):6-7. doi: 10.1093/cvr/cvv039. Epub 2015 Feb 17. Cardiovasc Res. 2015. PMID: 25691542 No abstract available.
-
Prolonged action potentials in HCM-derived iPSC--biology or artefact?Cardiovasc Res. 2015 Apr 1;106(1):6. doi: 10.1093/cvr/cvv038. Epub 2015 Feb 17. Cardiovasc Res. 2015. PMID: 25691543 No abstract available.
References
-
- Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. - PubMed
-
- Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. - PubMed
-
- Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;60:705–715. - PubMed
-
- Coppini R, Ferrantini C, Yao L, Fan P, Del Lungo M, Stillitano F, Sartiani L, Tosi B, Suffredini S, Tesi C, Yacoub M, Olivotto I, Belardinelli L, Poggesi C, Cerbai E, Mugelli A. Late sodium current inhibition reverses electromechanical dysfunction in human hypertrophic cardiomyopathy. Circulation. 2013;127:575–584. - PubMed
-
- Tomaselli GF, Marban E. Electrophysiological remodeling in hypertrophy and heart failure. Cardiovasc Res. 1999;42:270–283. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

