A prominent and conserved role for YY1 in Xist transcriptional activation
- PMID: 25209548
- PMCID: PMC4172967
- DOI: 10.1038/ncomms5878
A prominent and conserved role for YY1 in Xist transcriptional activation
Abstract
Accumulation of the noncoding RNA Xist on one X chromosome in female cells is a hallmark of X-chromosome inactivation (XCI) in eutherians. Here we uncover an essential function for the ubiquitous autosomal transcription factor Yin-Yang 1 (YY1) in the transcriptional activation of Xist in both human and mouse. We show that loss of YY1 prevents Xist upregulation during the initiation and maintenance of X-inactivation, and that YY1 binds directly the Xist 5' region to trigger the activity of the Xist promoter. Binding of YY1 to the Xist 5' region before XCI competes with the Xist repressor REX1, whereas DNA methylation controls mono-allelic fixation of YY1 to Xist at the onset of XCI. YY1 is thus the first autosomal activating factor involved in a fundamental and conserved pathway of Xist regulation that ensures the asymmetric transcriptional upregulation of the master regulator of XCI.
Figures

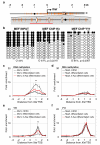


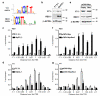


References
-
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–3. - PubMed
-
- Brockdorff N, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–31. - PubMed
-
- Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. - PubMed
-
- Borsani G, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–9. - PubMed
-
- Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12:429–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

