Gibbon genome and the fast karyotype evolution of small apes
- PMID: 25209798
- PMCID: PMC4249732
- DOI: 10.1038/nature13679
Gibbon genome and the fast karyotype evolution of small apes
Abstract
Gibbons are small arboreal apes that display an accelerated rate of evolutionary chromosomal rearrangement and occupy a key node in the primate phylogeny between Old World monkeys and great apes. Here we present the assembly and analysis of a northern white-cheeked gibbon (Nomascus leucogenys) genome. We describe the propensity for a gibbon-specific retrotransposon (LAVA) to insert into chromosome segregation genes and alter transcription by providing a premature termination site, suggesting a possible molecular mechanism for the genome plasticity of the gibbon lineage. We further show that the gibbon genera (Nomascus, Hylobates, Hoolock and Symphalangus) experienced a near-instantaneous radiation ∼5 million years ago, coincident with major geographical changes in southeast Asia that caused cycles of habitat compression and expansion. Finally, we identify signatures of positive selection in genes important for forelimb development (TBX5) and connective tissues (COL1A1) that may have been involved in the adaptation of gibbons to their arboreal habitat.
Conflict of interest statement
E.E.E. is on the scientific advisory board (SAB) of DNAnexus and was an SAB member of Pacific Biosciences (2009–2013) and SynapDx (2011–2013).
Figures




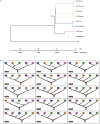
Comment in
-
Genomics: Something to swing about.Nature. 2014 Sep 11;513(7517):174-5. doi: 10.1038/513174a. Nature. 2014. PMID: 25209792 No abstract available.
References
-
- Mittermeier, R. A., Rylands, A. B. & Wilson, D. E. Handbook of the Mammals of the World Vol. 3 (Lynx Edicions,. (2013)
-
- Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- SRA/SRP043117
Grants and funding
- R01 MH081203/MH/NIMH NIH HHS/United States
- P51 RR000163/RR/NCRR NIH HHS/United States
- U41 HG007497/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- U41 HG007234/HG/NHGRI NIH HHS/United States
- R01 HG002939/HG/NHGRI NIH HHS/United States
- R01 GM59290/GM/NIGMS NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- T15 LM007088/LM/NLM NIH HHS/United States
- R01 HG002385/HG/NHGRI NIH HHS/United States
- WT098051/WT_/Wellcome Trust/United Kingdom
- U41 HG007497-01/HG/NHGRI NIH HHS/United States
- U54HG003273/HG/NHGRI NIH HHS/United States
- R01 HG005226/HG/NHGRI NIH HHS/United States
- 260372/ERC_/European Research Council/International
- P30CA006973/CA/NCI NIH HHS/United States
- P30 CA006973/CA/NCI NIH HHS/United States
- P30 AA019355/AA/NIAAA NIH HHS/United States
- HG002385/HG/NHGRI NIH HHS/United States
- R01 GM059290/GM/NIGMS NIH HHS/United States
- 095908/WT_/Wellcome Trust/United Kingdom
- 15603/CRUK_/Cancer Research UK/United Kingdom
- U41HG007234/HG/NHGRI NIH HHS/United States
- R01_HG005226/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

