Absence of SUN-domain protein Slp1 blocks karyogamy and switches meiotic recombination and synapsis from homologs to sister chromatids
- PMID: 25210014
- PMCID: PMC4183305
- DOI: 10.1073/pnas.1415758111
Absence of SUN-domain protein Slp1 blocks karyogamy and switches meiotic recombination and synapsis from homologs to sister chromatids
Abstract
Karyogamy, the process of nuclear fusion is required for two haploid gamete nuclei to form a zygote. Also, in haplobiontic organisms, karyogamy is required to produce the diploid nucleus/cell that then enters meiosis. We identify sun like protein 1 (Slp1), member of the mid-Sad1p, UNC-84-domain ubiquitous family, as essential for karyogamy in the filamentous fungus Sordaria macrospora, thus uncovering a new function for this protein family. Slp1 is required at the last step, nuclear fusion, not for earlier events including nuclear movements, recognition, and juxtaposition. Correspondingly, like other family members, Slp1 localizes to the endoplasmic reticulum and also to its extensions comprising the nuclear envelope. Remarkably, despite the absence of nuclear fusion in the slp1 null mutant, meiosis proceeds efficiently in the two haploid "twin" nuclei, by the same program and timing as in diploid nuclei with a single dramatic exception: the normal prophase program of recombination and synapsis between homologous chromosomes, including loading of recombination and synaptonemal complex proteins, occurs instead between sister chromatids. Moreover, the numbers of recombination-initiating double-strand breaks (DSBs) and ensuing recombinational interactions, including foci of the essential crossover factor Homo sapiens enhancer of invasion 10 (Hei10), occur at half the diploid level in each haploid nucleus, implying per-chromosome specification of DSB formation. Further, the distribution of Hei10 foci shows interference like in diploid meiosis. Centromere and spindle dynamics, however, still occur in the diploid mode during the two meiotic divisions. These observations imply that the prophase program senses absence of karyogamy and/or absence of a homolog partner and adjusts the interchromosomal interaction program accordingly.
Keywords: recombination/synapsis; sisters versus homologs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

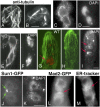
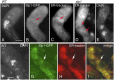
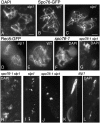

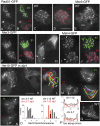
Similar articles
-
Asy2/Mer2: an evolutionarily conserved mediator of meiotic recombination, pairing, and global chromosome compaction.Genes Dev. 2017 Sep 15;31(18):1880-1893. doi: 10.1101/gad.304543.117. Epub 2017 Oct 11. Genes Dev. 2017. PMID: 29021238 Free PMC article.
-
Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis.J Cell Biol. 2009 Sep 7;186(5):713-25. doi: 10.1083/jcb.200810107. J Cell Biol. 2009. PMID: 19736318 Free PMC article.
-
Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition.Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12865-70. doi: 10.1073/pnas.2034282100. Epub 2003 Oct 16. Proc Natl Acad Sci U S A. 2003. PMID: 14563920 Free PMC article.
-
Sordaria, a model system to uncover links between meiotic pairing and recombination.Semin Cell Dev Biol. 2016 Jun;54:149-57. doi: 10.1016/j.semcdb.2016.02.012. Epub 2016 Feb 10. Semin Cell Dev Biol. 2016. PMID: 26877138 Free PMC article. Review.
-
Prelude to a division.Annu Rev Cell Dev Biol. 2008;24:397-424. doi: 10.1146/annurev.cellbio.23.090506.123245. Annu Rev Cell Dev Biol. 2008. PMID: 18597662 Free PMC article. Review.
Cited by
-
Control of Meiotic Crossovers: From Double-Strand Break Formation to Designation.Annu Rev Genet. 2016 Nov 23;50:175-210. doi: 10.1146/annurev-genet-120215-035111. Epub 2016 Sep 14. Annu Rev Genet. 2016. PMID: 27648641 Free PMC article. Review.
-
RNAi-Related Dicer and Argonaute Proteins Play Critical Roles for Meiocyte Formation, Chromosome-Axes Lengths and Crossover Patterning in the Fungus Sordaria macrospora.Front Cell Dev Biol. 2021 Jun 28;9:684108. doi: 10.3389/fcell.2021.684108. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34262901 Free PMC article.
-
In Depth Topological Analysis of Arabidopsis Mid-SUN Proteins and Their Interaction with the Membrane-Bound Transcription Factor MaMYB.Plants (Basel). 2023 Apr 27;12(9):1787. doi: 10.3390/plants12091787. Plants (Basel). 2023. PMID: 37176845 Free PMC article.
-
Canonical and noncanonical roles of Hop1 are crucial for meiotic prophase in the fungus Sordaria macrospora.PLoS Biol. 2024 Jul 1;22(7):e3002705. doi: 10.1371/journal.pbio.3002705. eCollection 2024 Jul. PLoS Biol. 2024. PMID: 38950075 Free PMC article.
-
Evolution and Functional Divergence of SUN Genes in Plants.Front Plant Sci. 2021 Mar 8;12:646622. doi: 10.3389/fpls.2021.646622. eCollection 2021. Front Plant Sci. 2021. PMID: 33763102 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

