Evolutionary aspects of enzyme dynamics
- PMID: 25210031
- PMCID: PMC4215204
- DOI: 10.1074/jbc.R114.565515
Evolutionary aspects of enzyme dynamics
Abstract
The role of evolutionary pressure on the chemical step catalyzed by enzymes is somewhat enigmatic, in part because chemistry is not rate-limiting for many optimized systems. Herein, we present studies that examine various aspects of the evolutionary relationship between protein dynamics and the chemical step in two paradigmatic enzyme families, dihydrofolate reductases and alcohol dehydrogenases. Molecular details of both convergent and divergent evolution are beginning to emerge. The findings suggest that protein dynamics across an entire enzyme can play a role in adaptation to differing physiological conditions. The growing tool kit of kinetics, kinetic isotope effects, molecular biology, biophysics, and bioinformatics provides means to link evolutionary changes in structure-dynamics function to the vibrational and conformational states of each protein.
Keywords: Alcohol Dehydrogenase (ADH); Enzyme; Enzyme Catalysis; Enzyme Kinetics; Enzyme Mechanism; Enzyme Mutation; Evolution; Folate Metabolism; Molecular Evolution; Protein Evolution.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
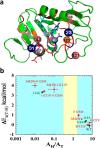
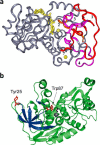
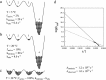
References
-
- Süel G. M., Lockless S. W., Wall M. A., Ranganathan R. (2003) Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 10, 59–69 - PubMed
-
- Hammes-Schiffer S., Benkovic S. J. (2006) Relating protein motion to catalysis. Annu. Rev. Biochem. 75, 519–541 - PubMed
-
- Radkiewicz J. L., Brooks C. L. (2000) Protein dynamics in enzymatic catalysis: exploration of dihydrofolate reductase. J. Am. Chem. Soc. 122, 225–231
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

