Ral GTPase down-regulation stabilizes and reactivates p53 to inhibit malignant transformation
- PMID: 25210032
- PMCID: PMC4223330
- DOI: 10.1074/jbc.M114.565796
Ral GTPase down-regulation stabilizes and reactivates p53 to inhibit malignant transformation
Abstract
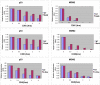
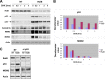
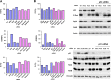
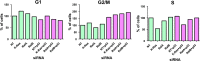
Ral GTPases are critical effectors of Ras, yet the molecular mechanism by which they induce malignant transformation is not well understood. In this study, we found the expression of K-Ras, RalB, and sometimes RalA, but not AKT1/2 and c-Raf, to be required for maintaining low levels of p53 in human cancer cells that harbor mutant K-Ras and wild-type p53. Down-regulation of K-Ras, RalB, and sometimes RalA increases p53 protein levels and results in a p53-dependent up-regulation of the expression of p21(WAF). K-Ras, RalA, and RalB depletion increases p53 stability as demonstrated by ataxia telangiectasia-mutated kinase activation, increased Ser-15 phosphorylation, and a significant (up to 6-fold) increase in p53 half-life. Furthermore, depletion of K-Ras and RalB inhibits anchorage-independent growth and invasion and interferes with cell cycle progression in a p53-dependent manner. Depletion of RalA inhibits invasion in a p53-dependent manner. Thus, expression of K-Ras and RalB and possibly RalA proteins is critical for maintaining low levels of p53, and down-regulation of these GTPases reactivates p53 by significantly enhancing its stability, and this contributes to suppression of malignant transformation.
Keywords: Apoptosis; Cancer Biology; K-Ras; RalA; RalB; Ras Protein; Serine 15 Phosphorylation; Signal Transduction; p53.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





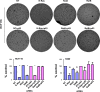
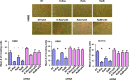
Similar articles
-
The GTPase KRAS suppresses the p53 tumor suppressor by activating the NRF2-regulated antioxidant defense system in cancer cells.J Biol Chem. 2020 Mar 6;295(10):3055-3063. doi: 10.1074/jbc.RA119.011930. Epub 2020 Jan 30. J Biol Chem. 2020. PMID: 32001619 Free PMC article.
-
Depletion of K-Ras promotes proteasome degradation of survivin.Cell Cycle. 2013 Feb 1;12(3):522-32. doi: 10.4161/cc.23407. Epub 2013 Jan 16. Cell Cycle. 2013. PMID: 23324341 Free PMC article.
-
Ras-related small GTPases RalA and RalB regulate cellular survival after ionizing radiation.Int J Radiat Oncol Biol Phys. 2010 Sep 1;78(1):205-12. doi: 10.1016/j.ijrobp.2010.03.023. Epub 2010 Jul 7. Int J Radiat Oncol Biol Phys. 2010. PMID: 20619549 Free PMC article.
-
Ral GTPases in tumorigenesis: emerging from the shadows.Exp Cell Res. 2013 Sep 10;319(15):2337-42. doi: 10.1016/j.yexcr.2013.06.020. Epub 2013 Jul 2. Exp Cell Res. 2013. PMID: 23830877 Free PMC article. Review.
-
The RAL Enigma: Distinct Roles of RALA and RALB in Cancer.Cells. 2022 May 14;11(10):1645. doi: 10.3390/cells11101645. Cells. 2022. PMID: 35626682 Free PMC article. Review.
Cited by
-
Controlled Plasma Membrane Delivery of FGFR1 and Modulation of Signaling by a Novel Regulated Anterograde RTK Transport Pathway.Cancers (Basel). 2023 Dec 14;15(24):5837. doi: 10.3390/cancers15245837. Cancers (Basel). 2023. PMID: 38136383 Free PMC article.
-
A genome-wide siRNA screen for regulators of tumor suppressor p53 activity in human non-small cell lung cancer cells identifies components of the RNA splicing machinery as targets for anticancer treatment.Mol Oncol. 2017 May;11(5):534-551. doi: 10.1002/1878-0261.12052. Epub 2017 Apr 11. Mol Oncol. 2017. PMID: 28296343 Free PMC article.
-
RALB GTPase: a critical regulator of DR5 expression and TRAIL sensitivity in KRAS mutant colorectal cancer.Cell Death Dis. 2020 Oct 29;11(10):930. doi: 10.1038/s41419-020-03131-3. Cell Death Dis. 2020. PMID: 33122623 Free PMC article.
-
SIRT2 and Lysine Fatty Acylation Regulate the Activity of RalB and Cell Migration.ACS Chem Biol. 2019 Sep 20;14(9):2014-2023. doi: 10.1021/acschembio.9b00492. Epub 2019 Sep 3. ACS Chem Biol. 2019. PMID: 31433161 Free PMC article.
-
Hypoxic resistance of KRAS mutant tumor cells to 3-Bromopyruvate is counteracted by Prima-1 and reversed by N-acetylcysteine.BMC Cancer. 2016 Nov 18;16(1):902. doi: 10.1186/s12885-016-2930-9. BMC Cancer. 2016. PMID: 27863474 Free PMC article.
References
-
- Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 - PubMed
-
- Forbes S. A., Bindal N., Bamford S., Cole C., Kok C. Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A., Teague J. W., Campbell P. J., Stratton M. R., Futreal P. A. (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39, D945–D950 - PMC - PubMed
-
- Downward J. (2003) Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11–22 - PubMed
-
- Brown C. J., Lain S., Verma C. S., Fersht A. R., Lane D. P. (2009) Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer 9, 862–873 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

