Hallmarks of hepatitis C virus in equine hepacivirus
- PMID: 25210167
- PMCID: PMC4249100
- DOI: 10.1128/JVI.02280-14
Hallmarks of hepatitis C virus in equine hepacivirus
Abstract
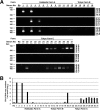
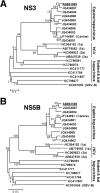
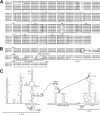
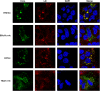
Equine hepacivirus (EHcV) has been identified as a closely related homologue of hepatitis C virus (HCV) in the United States, the United Kingdom, and Germany, but not in Asian countries. In this study, we genetically and serologically screened 31 serum samples obtained from Japanese-born domestic horses for EHcV infection and subsequently identified 11 PCR-positive and 7 seropositive serum samples. We determined the full sequence of the EHcV genome, including the 3' untranslated region (UTR), which had previously not been completely revealed. The polyprotein of a Japanese EHcV strain showed approximately 95% homology to those of the reported strains. HCV-like cis-acting RNA elements, including the stem-loop structures of the 3' UTR and kissing-loop interaction were deduced from regions around both UTRs of the EHcV genome. A comparison of the EHcV and HCV core proteins revealed that Ile(190) and Phe(191) of the EHcV core protein could be important for cleavage of the core protein by signal peptide peptidase (SPP) and were replaced with Ala and Leu, respectively, which inhibited intramembrane cleavage of the EHcV core protein. The loss-of-function mutant of SPP abrogated intramembrane cleavage of the EHcV core protein and bound EHcV core protein, suggesting that the EHcV core protein may be cleaved by SPP to become a mature form. The wild-type EHcV core protein, but not the SPP-resistant mutant, was localized on lipid droplets and partially on the lipid raft-like membrane in a manner similar to that of the HCV core protein. These results suggest that EHcV may conserve the genetic and biological properties of HCV.
Importance: EHcV, which shows the highest amino acid or nucleotide homology to HCV among hepaciviruses, was previously reported to infect horses from Western, but not Asian, countries. We herein report EHcV infection in Japanese-born horses. In this study, HCV-like RNA secondary structures around both UTRs were predicted by determining the whole-genome sequence of EHcV. Our results also suggest that the EHcV core protein is cleaved by SPP to become a mature form and then is localized on lipid droplets and partially on lipid raft-like membranes in a manner similar to that of the HCV core protein. Hence, EHcV was identified as a closely related homologue of HCV based on its genetic structure as well as its biological properties. A clearer understanding of the epidemiology, genetic structure, and infection mechanism of EHcV will assist in elucidating the evolution of hepaciviruses as well as the development of surrogate models for the study of HCV.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ, Ellison JA, Gilbert AT, Kuzmin IV, Niezgoda M, Osinubi MO, Recuenco S, Markotter W, Breiman RF, Kalemba L, Malekani J, Lindblade KA, Rostal MK, Ojeda-Flores R, Suzan G, Davis LB, Blau DM, Ogunkoya AB, Alvarez Castillo DA, Moran D, Ngam S, Akaibe D, Agwanda B, Briese T, Epstein JH, Daszak P, Rupprecht CE, Holmes EC, Lipkin WI. 2013. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. U. S. A. 110:8194–8199. 10.1073/pnas.1303037110. - DOI - PMC - PubMed
-
- Drexler JF, Corman VM, Muller MA, Lukashev AN, Gmyl A, Coutard B, Adam A, Ritz D, Leijten LM, van Riel D, Kallies R, Klose SM, Gloza-Rausch F, Binger T, Annan A, Adu-Sarkodie Y, Oppong S, Bourgarel M, Rupp D, Hoffmann B, Schlegel M, Kummerer BM, Kruger DH, Schmidt-Chanasit J, Setien AA, Cottontail VM, Hemachudha T, Wacharapluesadee S, Osterrieder K, Bartenschlager R, Matthee S, Beer M, Kuiken T, Reusken C, Leroy EM, Ulrich RG, Drosten C. 2013. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 9:e1003438. 10.1371/journal.ppat.1003438. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

