Cholesterol-dependent membrane fusion induced by the gp41 membrane-proximal external region-transmembrane domain connection suggests a mechanism for broad HIV-1 neutralization
- PMID: 25210180
- PMCID: PMC4249078
- DOI: 10.1128/JVI.02151-14
Cholesterol-dependent membrane fusion induced by the gp41 membrane-proximal external region-transmembrane domain connection suggests a mechanism for broad HIV-1 neutralization
Abstract
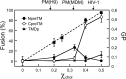
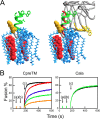
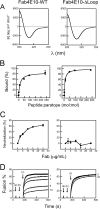
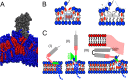
The HIV-1 glycoprotein 41 promotes fusion of the viral membrane with that of the target cell. Structural, biochemical, and biophysical studies suggest that its membrane-proximal external region (MPER) may interact with the HIV-1 membrane and induce its disruption and/or deformation during the process. However, the high cholesterol content of the envelope (ca. 40 to 50 mol%) imparts high rigidity, thereby acting against lipid bilayer restructuring. Here, based on the outcome of vesicle stability assays, all-atom molecular dynamics simulations, and atomic force microscopy observations, we propose that the conserved sequence connecting the MPER with the N-terminal residues of the transmembrane domain (TMD) is involved in HIV-1 fusion. This junction would function by inducing phospholipid protrusion and acyl-chain splay in the cholesterol-enriched rigid envelope. Supporting the functional relevance of such a mechanism, membrane fusion was inhibited by the broadly neutralizing 4E10 antibody but not by a nonneutralizing variant with the CDR-H3 loop deleted. We conclude that the MPER-TMD junction embodies an envelope-disrupting C-terminal fusion peptide that can be targeted by broadly neutralizing antibodies.
Importance: Fusion of the cholesterol-enriched viral envelope with the cell membrane marks the beginning of the infectious HIV-1 replicative cycle. Consequently, the Env glycoprotein-mediated fusion function constitutes an important clinical target for inhibitors and preventive vaccines. Antibodies 4E10 and 10E8 bind to one Env vulnerability site located at the gp41 membrane-proximal external region (MPER)-transmembrane domain (TMD) junction and block infection. These antibodies display broad viral neutralization, which underscores the conservation and functionality of the MPER-TMD region. In this work, we combined biochemical assays with molecular dynamics simulations and microscopy observations to characterize the unprecedented fusogenic activity of the MPER-TMD junction. The fact that such activity is dependent on cholesterol and inhibited by the broadly neutralizing 4E10 antibody emphasizes its physiological relevance. Discovery of this functional element adds to our understanding of the mechanisms underlying HIV-1 infection and its blocking by antibodies.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

