The HERV-K human endogenous retrovirus envelope protein antagonizes Tetherin antiviral activity
- PMID: 25210194
- PMCID: PMC4248984
- DOI: 10.1128/JVI.02234-14
The HERV-K human endogenous retrovirus envelope protein antagonizes Tetherin antiviral activity
Abstract
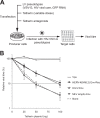
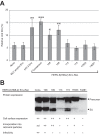
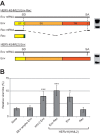
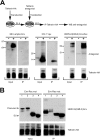
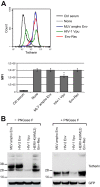
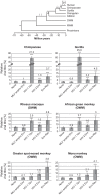
Endogenous retroviruses are the remnants of past retroviral infections that are scattered within mammalian genomes. In humans, most of these elements are old degenerate sequences that have lost their coding properties. The HERV-K(HML2) family is an exception: it recently amplified in the human genome and corresponds to the most active proviruses, with some intact open reading frames and the potential to encode viral particles. Here, using a reconstructed consensus element, we show that HERV-K(HML2) proviruses are able to inhibit Tetherin, a cellular restriction factor that is active against most enveloped viruses and acts by keeping the viral particles attached to the cell surface. More precisely, we identify the Envelope protein (Env) as the viral effector active against Tetherin. Through immunoprecipitation experiments, we show that the recognition of Tetherin is mediated by the surface subunit of Env. Similar to Ebola glycoprotein, HERV-K(HML2) Env does not mediate Tetherin degradation or cell surface removal; therefore, it uses a yet-undescribed mechanism to inactivate Tetherin. We also assessed all natural complete alleles of endogenous HERV-K(HML2) Env described to date for their ability to inhibit Tetherin and found that two of them (out of six) can block Tetherin restriction. However, due to their recent amplification, HERV-K(HML2) elements are extremely polymorphic in the human population, and it is likely that individuals will not all possess the same anti-Tetherin potential. Because of Tetherin's role as a restriction factor capable of inducing innate immune responses, this could have functional consequences for individual responses to infection.
Importance: Tetherin, a cellular protein initially characterized for its role against HIV-1, has been proven to counteract numerous enveloped viruses. It blocks the release of viral particles from producer cells, keeping them tethered to the cell surface. Several viruses have developed strategies to inhibit Tetherin activity, allowing them to efficiently infect and replicate in their host. Here, we show that human HERV-K(HML2) elements, the remnants of an ancient retroviral infection, possess an anti-Tetherin activity which is mediated by the envelope protein. It is likely that this activity was an important factor that contributed to the recent, human-specific amplification of this family of elements. Also, due to their recent amplification, HERV-K(HML2) elements are highly polymorphic in the human population. Since Tetherin is a mediator of innate immunity, interindividual variations among HERV-K(HML2) Env genes may result in differences in immune responses to infection.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321–3329. 10.1128/JVI.74.7.3321-3329.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

