Neighbor preferences of amino acids and context-dependent effects of amino acid substitutions in human, mouse, and dog
- PMID: 25210846
- PMCID: PMC4200849
- DOI: 10.3390/ijms150915963
Neighbor preferences of amino acids and context-dependent effects of amino acid substitutions in human, mouse, and dog
Abstract
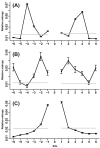
Amino acids show apparent propensities toward their neighbors. In addition to preferences of amino acids for their neighborhood context, amino acid substitutions are also considered to be context-dependent. However, context-dependence patterns of amino acid substitutions still remain poorly understood. Using relative entropy, we investigated the neighbor preferences of 20 amino acids and the context-dependent effects of amino acid substitutions with protein sequences in human, mouse, and dog. For 20 amino acids, the highest relative entropy was mostly observed at the nearest adjacent site of either N- or C-terminus except C and G. C showed the highest relative entropy at the third flanking site and periodic pattern was detected at G flanking sites. Furthermore, neighbor preference patterns of amino acids varied greatly in different secondary structures. We then comprehensively investigated the context-dependent effects of amino acid substitutions. Our results showed that nearly half of 380 substitution types were evidently context dependent, and the context-dependent patterns relied on protein secondary structures. Among 20 amino acids, P elicited the greatest effect on amino acid substitutions. The underlying mechanisms of context-dependent effects of amino acid substitutions were possibly mutation bias at a DNA level and natural selection. Our findings may improve secondary structure prediction algorithms and protein design; moreover, this study provided useful information to develop empirical models of protein evolution that consider dependence between residues.
Figures
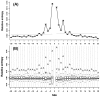
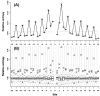
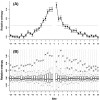
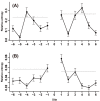
 at each flanking site of the amino acid A. The bold line in each box represents the median of the 20 values. The top and bottom lines of each box indicate the upper and lower quartiles, respectively. The upper and lower whiskers represent the largest data point which was less than the sum of the upper quartile plus 1.5 times the interquartile range (IQR), and the lowest data point which was greater than the lower quartile minus 1.5 IQR, respectively. In order to determine which amino acids were apparently preferred (or not preferred) in the neighboring sites, the outliers were represented by the corresponding one letter codes of amino acids.
at each flanking site of the amino acid A. The bold line in each box represents the median of the 20 values. The top and bottom lines of each box indicate the upper and lower quartiles, respectively. The upper and lower whiskers represent the largest data point which was less than the sum of the upper quartile plus 1.5 times the interquartile range (IQR), and the lowest data point which was greater than the lower quartile minus 1.5 IQR, respectively. In order to determine which amino acids were apparently preferred (or not preferred) in the neighboring sites, the outliers were represented by the corresponding one letter codes of amino acids.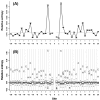
Similar articles
-
Protein structure, neighbor effect, and a new index of amino acid dissimilarities.Mol Biol Evol. 2002 Jan;19(1):58-67. doi: 10.1093/oxfordjournals.molbev.a003982. Mol Biol Evol. 2002. PMID: 11752190
-
Effects of side-chain characteristics on stability and oligomerization state of a de novo-designed model coiled-coil: 20 amino acid substitutions in position "d".J Mol Biol. 2000 Jul 7;300(2):377-402. doi: 10.1006/jmbi.2000.3866. J Mol Biol. 2000. PMID: 10873472
-
Residual dipolar couplings in short peptides reveal systematic conformational preferences of individual amino acids.J Am Chem Soc. 2006 Oct 18;128(41):13508-14. doi: 10.1021/ja063606h. J Am Chem Soc. 2006. PMID: 17031964
-
Correlated substitution analysis and the prediction of amino acid structural contacts.Brief Bioinform. 2008 Jan;9(1):46-56. doi: 10.1093/bib/bbm052. Epub 2007 Nov 13. Brief Bioinform. 2008. PMID: 18000015 Review.
-
Prediction of contacts from correlated sequence substitutions.Curr Opin Struct Biol. 2013 Jun;23(3):473-9. doi: 10.1016/j.sbi.2013.04.001. Epub 2013 May 14. Curr Opin Struct Biol. 2013. PMID: 23680395 Review.
Cited by
-
Distribution of dipeptides in different protein structural classes: an effort to find new similarities.Eur Biophys J. 2018 Jan;47(1):31-38. doi: 10.1007/s00249-017-1226-6. Epub 2017 Jun 13. Eur Biophys J. 2018. PMID: 28612124
-
Convolutional Neural Network Based Approach to in Silico Non-Anticipating Prediction of Antigenic Distance for Influenza Virus.Viruses. 2020 Sep 12;12(9):1019. doi: 10.3390/v12091019. Viruses. 2020. PMID: 32932748 Free PMC article.
-
Chameleon sequences in neurodegenerative diseases.Biochem Biophys Res Commun. 2016 Mar 25;472(1):209-16. doi: 10.1016/j.bbrc.2016.01.187. Epub 2016 Feb 23. Biochem Biophys Res Commun. 2016. PMID: 26920059 Free PMC article.
-
A reactive center loop-based prediction platform to enhance the design of therapeutic SERPINs.Proc Natl Acad Sci U S A. 2021 Nov 9;118(45):e2108458118. doi: 10.1073/pnas.2108458118. Proc Natl Acad Sci U S A. 2021. PMID: 34740972 Free PMC article.
References
-
- Chou P.Y. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978;47:45–148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

