Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae β-galactosidase, BgaA
- PMID: 25210925
- PMCID: PMC4161441
- DOI: 10.1371/journal.ppat.1004364
Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae β-galactosidase, BgaA
Abstract
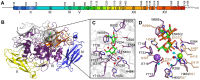
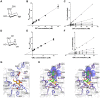
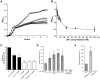
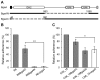
Bacterial cell-surface proteins play integral roles in host-pathogen interactions. These proteins are often architecturally and functionally sophisticated and yet few studies of such proteins involved in host-pathogen interactions have defined the domains or modules required for specific functions. Streptococcus pneumoniae (pneumococcus), an opportunistic pathogen that is a leading cause of community acquired pneumonia, otitis media and bacteremia, is decorated with many complex surface proteins. These include β-galactosidase BgaA, which is specific for terminal galactose residues β-1-4 linked to glucose or N-acetylglucosamine and known to play a role in pneumococcal growth, resistance to opsonophagocytic killing, and adherence. This study defines the domains and modules of BgaA that are required for these distinct contributions to pneumococcal pathogenesis. Inhibitors of β-galactosidase activity reduced pneumococcal growth and increased opsonophagocytic killing in a BgaA dependent manner, indicating these functions require BgaA enzymatic activity. In contrast, inhibitors increased pneumococcal adherence suggesting that BgaA bound a substrate of the enzyme through a distinct module or domain. Extensive biochemical, structural and cell based studies revealed two newly identified non-enzymatic carbohydrate-binding modules (CBMs) mediate adherence to the host cell surface displayed lactose or N-acetyllactosamine. This finding is important to pneumococcal biology as it is the first adhesin-carbohydrate receptor pair identified, supporting the widely held belief that initial pneumococcal attachment is to a glycoconjugate. Perhaps more importantly, this is the first demonstration that a CBM within a carbohydrate-active enzyme can mediate adherence to host cells and thus this study identifies a new class of carbohydrate-binding adhesins and extends the paradigm of CBM function. As other bacterial species express surface-associated carbohydrate-active enzymes containing CBMs these findings have broad implications for bacterial adherence. Together, these data illustrate that comprehending the architectural sophistication of surface-attached proteins can increase our understanding of the different mechanisms by which these proteins can contribute to bacterial pathogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
BgaA acts as an adhesin to mediate attachment of some pneumococcal strains to human epithelial cells.Microbiology (Reading). 2011 Aug;157(Pt 8):2369-2381. doi: 10.1099/mic.0.045609-0. Epub 2011 May 20. Microbiology (Reading). 2011. PMID: 21602213 Free PMC article.
-
Pneumococcal BgaA Promotes Host Organ Bleeding and Coagulation in a Mouse Sepsis Model.Front Cell Infect Microbiol. 2022 Jul 1;12:844000. doi: 10.3389/fcimb.2022.844000. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35846740 Free PMC article.
-
CcpA-dependent and -independent control of beta-galactosidase expression in Streptococcus pneumoniae occurs via regulation of an upstream phosphotransferase system-encoding operon.J Bacteriol. 2007 Jul;189(14):5183-92. doi: 10.1128/JB.00449-07. Epub 2007 May 11. J Bacteriol. 2007. PMID: 17496092 Free PMC article.
-
Glycan-metabolizing enzymes in microbe-host interactions: the Streptococcus pneumoniae paradigm.FEBS Lett. 2018 Dec;592(23):3865-3897. doi: 10.1002/1873-3468.13045. Epub 2018 Apr 19. FEBS Lett. 2018. PMID: 29608212 Review.
-
Role of Streptococcus pneumoniae extracellular glycosidases in immune evasion.Front Cell Infect Microbiol. 2023 Feb 3;13:1109449. doi: 10.3389/fcimb.2023.1109449. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36816580 Free PMC article. Review.
Cited by
-
Role of BgaA as a Pneumococcal Virulence Factor Elucidated by Molecular Evolutionary Analysis.Front Microbiol. 2020 Sep 24;11:582437. doi: 10.3389/fmicb.2020.582437. eCollection 2020. Front Microbiol. 2020. PMID: 33072054 Free PMC article.
-
Within-host microevolution of Streptococcus pneumoniae is rapid and adaptive during natural colonisation.Nat Commun. 2020 Jul 10;11(1):3442. doi: 10.1038/s41467-020-17327-w. Nat Commun. 2020. PMID: 32651390 Free PMC article.
-
Role of Neuraminidase-Producing Bacteria in Exposing Cryptic Carbohydrate Receptors for Streptococcus gordonii Adherence.Infect Immun. 2018 Jun 21;86(7):e00068-18. doi: 10.1128/IAI.00068-18. Print 2018 Jul. Infect Immun. 2018. PMID: 29661931 Free PMC article.
-
Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.Nat Commun. 2018 Mar 13;9(1):1043. doi: 10.1038/s41467-018-03366-x. Nat Commun. 2018. PMID: 29535379 Free PMC article.
-
Emerging gut microbial glycoside hydrolase inhibitors.RSC Chem Biol. 2025 Jun 11;6(8):1233-1251. doi: 10.1039/d5cb00050e. eCollection 2025 Jul 30. RSC Chem Biol. 2025. PMID: 40534733 Free PMC article. Review.
References
-
- Ficko-Blean E, Boraston AB (2012) Insights into the recognition of the human glycome by microbial carbohydrate-binding modules. Curr Opin Struct Biol 22: 570–577. - PubMed
-
- King SJ, Hippe KR, Weiser JN (2006) Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae . Mol Microbiol 59: 961–974. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

