Regulation of arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression
- PMID: 25211338
- PMCID: PMC4161306
- DOI: 10.1371/journal.pgen.1004617
Regulation of arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression
Abstract
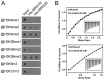
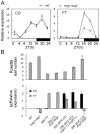
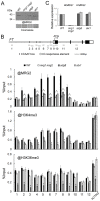
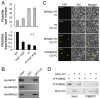
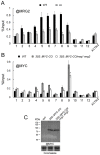
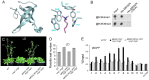
Day-length is important for regulating the transition to reproductive development (flowering) in plants. In the model plant Arabidopsis thaliana, the transcription factor CONSTANS (CO) promotes expression of the florigen FLOWERING LOCUS T (FT), constituting a key flowering pathway under long-day photoperiods. Recent studies have revealed that FT expression is regulated by changes of histone modification marks of the FT chromatin, but the epigenetic regulators that directly interact with the CO protein have not been identified. Here, we show that the Arabidopsis Morf Related Gene (MRG) group proteins MRG1 and MRG2 act as H3K4me3/H3K36me3 readers and physically interact with CO to activate FT expression. In vitro binding analyses indicated that the chromodomains of MRG1 and MRG2 preferentially bind H3K4me3/H3K36me3 peptides. The mrg1 mrg2 double mutant exhibits reduced mRNA levels of FT, but not of CO, and shows a late-flowering phenotype under the long-day but not short-day photoperiod growth conditions. MRG2 associates with the chromatin of FT promoter in a way dependent of both CO and H3K4me3/H3K36me3. Vice versa, loss of MRG1 and MRG2 also impairs CO binding at the FT promoter. Crystal structure analyses of MRG2 bound with H3K4me3/H3K36me3 peptides together with mutagenesis analysis in planta further demonstrated that MRG2 function relies on its H3K4me3/H3K36me3-binding activity. Collectively, our results unravel a novel chromatin regulatory mechanism, linking functions of MRG1 and MRG2 proteins, H3K4/H3K36 methylations, and CO in FT activation in the photoperiodic regulation of flowering time in plants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Amasino R (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013. - PubMed
-
- Ietswaart R, Wu Z, Dean C (2012) Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet 28: 445–453. - PubMed
-
- Andres F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639. - PubMed
-
- Turck F, Fornara F, Coupland G (2008) Regulation and Identity of Florigen: FLOWERING LOCUS T Moves Center Stage. Annual Review of Plant Biology 59: 573–594. - PubMed
-
- Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187: 57–66. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

