N-Methyl-4-hydrazino-7-nitrobenzofurazan: a fluorogenic substrate for peroxidase-like DNAzyme, and its potential application
- PMID: 25213215
- PMCID: PMC4206775
- DOI: 10.1007/s00216-014-8119-7
N-Methyl-4-hydrazino-7-nitrobenzofurazan: a fluorogenic substrate for peroxidase-like DNAzyme, and its potential application
Abstract
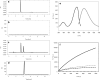
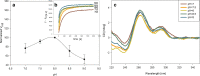
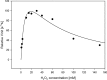
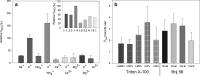
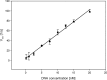
Characterization and optimization studies of N-methyl-4-hydrazino-7-nitrobenzofurazan (MNBDH) as a new fluorogenic substrate in the peroxidation reaction catalyzed by DNAzyme are reported. The effects of pH, H2O2 concentration, metal-cation type, and the concentration and type of surfactant on the fluorescence intensity were investigated. The optimized reaction was subsequently used for the development of an assay for DNA detection based on a molecular-beacon probe. The use of a fluorogenic substrate enabled the detection of a single-stranded DNA target with a 1 nmol L(-1) detection limit.
Figures


Similar articles
-
A simple mix-and-read bacteria detection system based on a DNAzyme and a molecular beacon.Chem Commun (Camb). 2019 Jun 20;55(51):7358-7361. doi: 10.1039/c9cc03746b. Chem Commun (Camb). 2019. PMID: 31172143
-
Fluorogenic substrate screening for G-quadruplex DNAzyme-based sensors.Biosens Bioelectron. 2013 Nov 15;49:312-7. doi: 10.1016/j.bios.2013.05.034. Epub 2013 May 29. Biosens Bioelectron. 2013. PMID: 23792624
-
A quencher-free DNAzyme beacon for fluorescently sensing uranyl ions via embedding 2-aminopurine.Biosens Bioelectron. 2019 Jun 15;135:166-172. doi: 10.1016/j.bios.2019.04.021. Epub 2019 Apr 15. Biosens Bioelectron. 2019. PMID: 31009884
-
DNA-nanoparticle micelles as supramolecular fluorogenic substrates enabling catalytic signal amplification and detection by DNAzyme probes.Chem Commun (Camb). 2011 Jan 7;47(1):167-9. doi: 10.1039/c0cc02291h. Epub 2010 Sep 7. Chem Commun (Camb). 2011. PMID: 20830351
-
Peroxidase-mimicking DNAzymes for biosensing applications: a review.Anal Chim Acta. 2011 Nov 30;707(1-2):7-17. doi: 10.1016/j.aca.2011.08.050. Epub 2011 Sep 16. Anal Chim Acta. 2011. PMID: 22027115 Review.
References
-
- Briones C, Martin-Gago JA. Nucleic Acids and Their Analogs as Nanomaterials for Biosensor Development. Curr Nanosci. 2006;2:257–273. doi: 10.2174/1573413710602030257. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

