Intersection of population variation and autoimmunity genetics in human T cell activation
- PMID: 25214635
- PMCID: PMC4751028
- DOI: 10.1126/science.1254665
Intersection of population variation and autoimmunity genetics in human T cell activation
Abstract
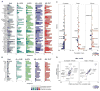
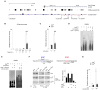
T lymphocyte activation by antigen conditions adaptive immune responses and immunopathologies, but we know little about its variation in humans and its genetic or environmental roots. We analyzed gene expression in CD4(+) T cells during unbiased activation or in T helper 17 (T(H)17) conditions from 348 healthy participants representing European, Asian, and African ancestries. We observed interindividual variability, most marked for cytokine transcripts, with clear biases on the basis of ancestry, and following patterns more complex than simple T(H)1/2/17 partitions. We identified 39 genetic loci specifically associated in cis with activated gene expression. We further fine-mapped and validated a single-base variant that modulates YY1 binding and the activity of an enhancer element controlling the autoimmune-associated IL2RA gene, affecting its activity in activated but not regulatory T cells. Thus, interindividual variability affects the fundamental immunologic process of T helper activation, with important connections to autoimmune disease.
Copyright © 2014, American Association for the Advancement of Science.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

