Phylogenetic information content of Copepoda ribosomal DNA repeat units: ITS1 and ITS2 impact
- PMID: 25215300
- PMCID: PMC4151598
- DOI: 10.1155/2014/926342
Phylogenetic information content of Copepoda ribosomal DNA repeat units: ITS1 and ITS2 impact
Abstract
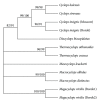
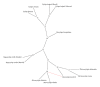
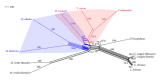
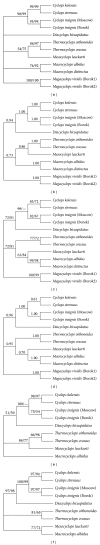
The utility of various regions of the ribosomal repeat unit for phylogenetic analysis was examined in 16 species representing four families, nine genera, and two orders of the subclass Copepoda (Crustacea). Fragments approximately 2000 bp in length containing the ribosomal DNA (rDNA) 18S and 28S gene fragments, the 5.8S gene, and the internal transcribed spacer regions I and II (ITS1 and ITS2) were amplified and analyzed. The DAMBE (Data Analysis in Molecular Biology and Evolution) software was used to analyze the saturation of nucleotide substitutions; this test revealed the suitability of both the 28S gene fragment and the ITS1/ITS2 rDNA regions for the reconstruction of phylogenetic trees. Distance (minimum evolution) and probabilistic (maximum likelihood, Bayesian) analyses of the data revealed that the 28S rDNA and the ITS1 and ITS2 regions are informative markers for inferring phylogenetic relationships among families of copepods and within the Cyclopidae family and associated genera. Split-graph analysis of concatenated ITS1/ITS2 rDNA regions of cyclopoid copepods suggested that the Mesocyclops, Thermocyclops, and Macrocyclops genera share complex evolutionary relationships. This study revealed that the ITS1 and ITS2 regions potentially represent different phylogenetic signals.
Figures
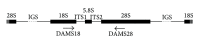

Similar articles
-
The ribosomal transcription units of Haplorchis pumilio and H. taichui and the use of 28S rDNA sequences for phylogenetic identification of common heterophyids in Vietnam.Parasit Vectors. 2017 Jan 9;10(1):17. doi: 10.1186/s13071-017-1968-0. Parasit Vectors. 2017. PMID: 28069063 Free PMC article.
-
Evolution and phylogenetic information content of the ribosomal DNA repeat unit in the Blattodea (Insecta).Insect Biochem Mol Biol. 2002 Sep;32(9):951-60. doi: 10.1016/s0965-1748(01)00164-3. Insect Biochem Mol Biol. 2002. PMID: 12213231
-
Phylogeny of freshwater parasitic copepods in the Ergasilidae (Copepoda: Poecilostomatoida) based on 18S and 28S rDNA sequences.Parasitol Res. 2008 Jan;102(2):299-306. doi: 10.1007/s00436-007-0764-8. Epub 2007 Oct 17. Parasitol Res. 2008. PMID: 17940799
-
Phylogenetic position of Creptotrema funduli in the Allocreadiidae based on partial 28S rDNA sequences.J Parasitol. 2012 Aug;98(4):873-5. doi: 10.1645/GE-3066.1. Epub 2012 Jan 30. J Parasitol. 2012. PMID: 22288517
-
Variation of rDNA Internal Transcribed Spacer Sequences in Rhizoctonia cerealis.Curr Microbiol. 2017 Jul;74(7):877-884. doi: 10.1007/s00284-017-1258-2. Epub 2017 May 5. Curr Microbiol. 2017. PMID: 28474105 Review.
Cited by
-
A synthesis tree of the Copepoda: integrating phylogenetic and taxonomic data reveals multiple origins of parasitism.PeerJ. 2021 Aug 18;9:e12034. doi: 10.7717/peerj.12034. eCollection 2021. PeerJ. 2021. PMID: 34466296 Free PMC article.
-
First Molecular Identification of Trypanosomes and Absence of Babesia sp. DNA in Faeces of Non-Human Primates in the Ecuadorian Amazon.Pathogens. 2022 Dec 7;11(12):1490. doi: 10.3390/pathogens11121490. Pathogens. 2022. PMID: 36558823 Free PMC article.
-
Phenotypic plasticity of life-history traits of a calanoid copepod in a tropical lake: Is the magnitude of thermal plasticity related to thermal variability?PLoS One. 2018 Apr 30;13(4):e0196496. doi: 10.1371/journal.pone.0196496. eCollection 2018. PLoS One. 2018. PMID: 29708999 Free PMC article.
-
Molecular phylogeny of Oncaeidae (Copepoda) using nuclear ribosomal internal transcribed spacer (ITS rDNA).PLoS One. 2017 Apr 25;12(4):e0175662. doi: 10.1371/journal.pone.0175662. eCollection 2017. PLoS One. 2017. PMID: 28441395 Free PMC article.
-
Chromatin diminution as a tool to study some biological problems.Comp Cytogenet. 2024 Feb 8;18:27-49. doi: 10.3897/compcytogen.17.112152. eCollection 2024. Comp Cytogenet. 2024. PMID: 38369988 Free PMC article.
References
-
- Boxshall GA, Defaye D. Global diversity of copepods (Crustacea: Copepoda) in freshwater. Hydrobiologia. 2008;595(1):195–207.
-
- Dussart B, Defaye D. World Directory of Crustacea Copepoda of Inland Waters. II—Cyclopiformes. Leiden, The Netherlands: Backhuys; 2006.
-
- Kiefer F. Versuch eines Systems der Cyclopiden. Zoologischer Anzeiger. 1927;73(11-12):302–308.
-
- Gurney R. British Fresh-Water Copepoda. III. Cyclopoida. London, UK: Ray Society; 1933.
-
- Rylov VM. Cyclopoida presnykh vod. Freshwater Cyclopoida V.3. Moscow, Russia: Leningrad; 1948.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

