Vfold: a web server for RNA structure and folding thermodynamics prediction
- PMID: 25215508
- PMCID: PMC4162592
- DOI: 10.1371/journal.pone.0107504
Vfold: a web server for RNA structure and folding thermodynamics prediction
Abstract
Background: The ever increasing discovery of non-coding RNAs leads to unprecedented demand for the accurate modeling of RNA folding, including the predictions of two-dimensional (base pair) and three-dimensional all-atom structures and folding stabilities. Accurate modeling of RNA structure and stability has far-reaching impact on our understanding of RNA functions in human health and our ability to design RNA-based therapeutic strategies.
Results: The Vfold server offers a web interface to predict (a) RNA two-dimensional structure from the nucleotide sequence, (b) three-dimensional structure from the two-dimensional structure and the sequence, and (c) folding thermodynamics (heat capacity melting curve) from the sequence. To predict the two-dimensional structure (base pairs), the server generates an ensemble of structures, including loop structures with the different intra-loop mismatches, and evaluates the free energies using the experimental parameters for the base stacks and the loop entropy parameters given by a coarse-grained RNA folding model (the Vfold model) for the loops. To predict the three-dimensional structure, the server assembles the motif scaffolds using structure templates extracted from the known PDB structures and refines the structure using all-atom energy minimization.
Conclusions: The Vfold-based web server provides a user friendly tool for the prediction of RNA structure and stability. The web server and the source codes are freely accessible for public use at "http://rna.physics.missouri.edu".
Conflict of interest statement
Figures


 in this example).
in this example).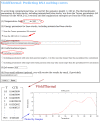

Similar articles
-
VfoldLA: A web server for loop assembly-based prediction of putative 3D RNA structures.J Struct Biol. 2019 Sep 1;207(3):235-240. doi: 10.1016/j.jsb.2019.06.002. Epub 2019 Jun 4. J Struct Biol. 2019. PMID: 31173857 Free PMC article.
-
cRNAsp12 Web Server for the Prediction of Circular RNA Secondary Structures and Stabilities.Int J Mol Sci. 2023 Feb 14;24(4):3822. doi: 10.3390/ijms24043822. Int J Mol Sci. 2023. PMID: 36835231 Free PMC article.
-
TBI server: a web server for predicting ion effects in RNA folding.PLoS One. 2015 Mar 23;10(3):e0119705. doi: 10.1371/journal.pone.0119705. eCollection 2015. PLoS One. 2015. PMID: 25798933 Free PMC article.
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
-
Theory and Modeling of RNA Structure and Interactions with Metal Ions and Small Molecules.Annu Rev Biophys. 2017 May 22;46:227-246. doi: 10.1146/annurev-biophys-070816-033920. Epub 2017 Mar 15. Annu Rev Biophys. 2017. PMID: 28301768 Free PMC article. Review.
Cited by
-
SimRNAweb: a web server for RNA 3D structure modeling with optional restraints.Nucleic Acids Res. 2016 Jul 8;44(W1):W315-9. doi: 10.1093/nar/gkw279. Epub 2016 Apr 19. Nucleic Acids Res. 2016. PMID: 27095203 Free PMC article.
-
Non-coding RNAs associated with Prader-Willi syndrome regulate transcription of neurodevelopmental genes in human induced pluripotent stem cells.Hum Mol Genet. 2023 Jan 27;32(4):608-620. doi: 10.1093/hmg/ddac228. Hum Mol Genet. 2023. PMID: 36084040 Free PMC article.
-
Modeling Structure, Stability, and Flexibility of Double-Stranded RNAs in Salt Solutions.Biophys J. 2018 Oct 16;115(8):1403-1416. doi: 10.1016/j.bpj.2018.08.030. Epub 2018 Aug 30. Biophys J. 2018. PMID: 30236782 Free PMC article.
-
Modeling Noncanonical RNA Base Pairs by a Coarse-Grained IsRNA2 Model.J Phys Chem B. 2021 Nov 4;125(43):11907-11915. doi: 10.1021/acs.jpcb.1c07288. Epub 2021 Oct 25. J Phys Chem B. 2021. PMID: 34694128 Free PMC article.
-
Terahertz waves regulate the mechanical unfolding of tau pre-mRNA hairpins.iScience. 2023 Aug 9;26(9):107572. doi: 10.1016/j.isci.2023.107572. eCollection 2023 Sep 15. iScience. 2023. PMID: 37664616 Free PMC article.
References
-
- Doudna JA, Cech TR (2002) The chemical repertoire of natural ribozymes. Nature 418: 222–228. - PubMed
-
- Bachellerie JP, Cavaille J, Huttenhofer A (2002) The expanding snoRNA world. Biochimie 84: 774–790. - PubMed
-
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat. Genet 39: 1278–1284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

