Lung fibroblasts from patients with idiopathic pulmonary fibrosis exhibit genome-wide differences in DNA methylation compared to fibroblasts from nonfibrotic lung
- PMID: 25215577
- PMCID: PMC4162578
- DOI: 10.1371/journal.pone.0107055
Lung fibroblasts from patients with idiopathic pulmonary fibrosis exhibit genome-wide differences in DNA methylation compared to fibroblasts from nonfibrotic lung
Abstract
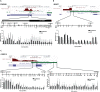
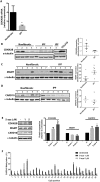
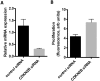
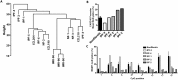
Excessive fibroproliferation is a central hallmark of idiopathic pulmonary fibrosis (IPF), a chronic, progressive disorder that results in impaired gas exchange and respiratory failure. Fibroblasts are the key effector cells in IPF, and aberrant expression of multiple genes contributes to their excessive fibroproliferative phenotype. DNA methylation changes are critical to the development of many diseases, but the DNA methylome of IPF fibroblasts has never been characterized. Here, we utilized the HumanMethylation 27 array, which assays the DNA methylation level of 27,568 CpG sites across the genome, to compare the DNA methylation patterns of IPF fibroblasts (n = 6) with those of nonfibrotic patient controls (n = 3) and commercially available normal lung fibroblast cell lines (n = 3). We found that multiple CpG sites across the genome are differentially methylated (as defined by P value less than 0.05 and fold change greater than 2) in IPF fibroblasts compared to fibroblasts from nonfibrotic controls. These methylation differences occurred both in genes recognized to be important in fibroproliferation and extracellular matrix generation, as well as in genes not previously recognized to participate in those processes (including organ morphogenesis and potassium ion channels). We used bisulfite sequencing to independently verify DNA methylation differences in 3 genes (CDKN2B, CARD10, and MGMT); these methylation changes corresponded with differences in gene expression at the mRNA and protein level. These differences in DNA methylation were stable throughout multiple cell passages. DNA methylation differences may thus help to explain a proportion of the differences in gene expression previously observed in studies of IPF fibroblasts. Moreover, significant variability in DNA methylation was observed among individual IPF cell lines, suggesting that differences in DNA methylation may contribute to fibroblast heterogeneity among patients with IPF. These results demonstrate that IPF fibroblasts exhibit global differences in DNA methylation that may contribute to the excessive fibroproliferation associated with this disease.
Conflict of interest statement
Figures

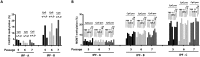
Similar articles
-
Global methylation patterns in idiopathic pulmonary fibrosis.PLoS One. 2012;7(4):e33770. doi: 10.1371/journal.pone.0033770. Epub 2012 Apr 10. PLoS One. 2012. PMID: 22506007 Free PMC article.
-
Hypermethylation-mediated silencing of p14(ARF) in fibroblasts from idiopathic pulmonary fibrosis.Am J Physiol Lung Cell Mol Physiol. 2012 Aug 15;303(4):L295-303. doi: 10.1152/ajplung.00332.2011. Epub 2012 Jun 15. Am J Physiol Lung Cell Mol Physiol. 2012. PMID: 22707614 Free PMC article.
-
Global DNA Methylation Pattern of Fibroblasts in Idiopathic Pulmonary Fibrosis.DNA Cell Biol. 2019 Sep;38(9):905-914. doi: 10.1089/dna.2018.4557. Epub 2019 Jul 15. DNA Cell Biol. 2019. PMID: 31305135
-
Epigenetics in lung fibrosis: from pathobiology to treatment perspective.Curr Opin Pulm Med. 2015 Sep;21(5):454-62. doi: 10.1097/MCP.0000000000000191. Curr Opin Pulm Med. 2015. PMID: 26176965 Free PMC article. Review.
-
Wilms Tumor 1-Driven Fibroblast Activation and Subpleural Thickening in Idiopathic Pulmonary Fibrosis.Int J Mol Sci. 2023 Feb 2;24(3):2850. doi: 10.3390/ijms24032850. Int J Mol Sci. 2023. PMID: 36769178 Free PMC article. Review.
Cited by
-
A Focus on "Eye on" Channels in Pulmonary Fibrosis.Am J Respir Cell Mol Biol. 2020 Feb;62(2):132-133. doi: 10.1165/rcmb.2019-0343ED. Am J Respir Cell Mol Biol. 2020. PMID: 31622111 Free PMC article. No abstract available.
-
Research Progress in the Molecular Mechanisms, Therapeutic Targets, and Drug Development of Idiopathic Pulmonary Fibrosis.Front Pharmacol. 2022 Jul 21;13:963054. doi: 10.3389/fphar.2022.963054. eCollection 2022. Front Pharmacol. 2022. PMID: 35935869 Free PMC article. Review.
-
Genome-wide analysis of DNA methylation and gene expression defines molecular characteristics of Crohn's disease-associated fibrosis.Clin Epigenetics. 2016 Mar 12;8:30. doi: 10.1186/s13148-016-0193-6. eCollection 2016. Clin Epigenetics. 2016. PMID: 26973718 Free PMC article.
-
Epigenetics Approaches toward Precision Medicine for Idiopathic Pulmonary Fibrosis: Focus on DNA Methylation.Biomedicines. 2023 Mar 28;11(4):1047. doi: 10.3390/biomedicines11041047. Biomedicines. 2023. PMID: 37189665 Free PMC article. Review.
-
Genome-Wide Epigenetic Signatures of Adaptive Developmental Plasticity in the Andes.Genome Biol Evol. 2021 Feb 3;13(2):evaa239. doi: 10.1093/gbe/evaa239. Genome Biol Evol. 2021. PMID: 33185669 Free PMC article.
References
-
- Bjoraker JA, Ryu JH, Edwin MK, Myers JL, Tazelaar HD, et al. (1998) Prognostic significance of histopathologic subsets in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 157: 199–203. - PubMed
-
- Jordana M, Schulman J, McSharry C, Irving LB, Newhouse MT, et al. (1988) Heterogeneous proliferative characteristics of human adult lung fibroblast lines and clonally derived fibroblasts from control and fibrotic tissue. Am Rev Respir Dis 137: 579–584. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

