Transcriptomics reveal several gene expression patterns in the piezophile Desulfovibrio hydrothermalis in response to hydrostatic pressure
- PMID: 25215865
- PMCID: PMC4162548
- DOI: 10.1371/journal.pone.0106831
Transcriptomics reveal several gene expression patterns in the piezophile Desulfovibrio hydrothermalis in response to hydrostatic pressure
Abstract
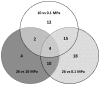
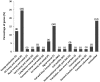
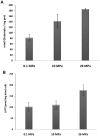
RNA-seq was used to study the response of Desulfovibrio hydrothermalis, isolated from a deep-sea hydrothermal chimney on the East-Pacific Rise at a depth of 2,600 m, to various hydrostatic pressure growth conditions. The transcriptomic datasets obtained after growth at 26, 10 and 0.1 MPa identified only 65 differentially expressed genes that were distributed among four main categories: aromatic amino acid and glutamate metabolisms, energy metabolism, signal transduction, and unknown function. The gene expression patterns suggest that D. hydrothermalis uses at least three different adaptation mechanisms, according to a hydrostatic pressure threshold (HPt) that was estimated to be above 10 MPa. Both glutamate and energy metabolism were found to play crucial roles in these mechanisms. Quantitation of the glutamate levels in cells revealed its accumulation at high hydrostatic pressure, suggesting its role as a piezolyte. ATP measurements showed that the energy metabolism of this bacterium is optimized for deep-sea life conditions. This study provides new insights into the molecular mechanisms linked to hydrostatic pressure adaptation in sulfate-reducing bacteria.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

