Comparative genome sequencing reveals genomic signature of extreme desiccation tolerance in the anhydrobiotic midge
- PMID: 25216354
- PMCID: PMC4175575
- DOI: 10.1038/ncomms5784
Comparative genome sequencing reveals genomic signature of extreme desiccation tolerance in the anhydrobiotic midge
Abstract
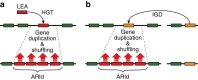
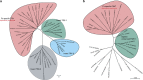
Anhydrobiosis represents an extreme example of tolerance adaptation to water loss, where an organism can survive in an ametabolic state until water returns. Here we report the first comparative analysis examining the genomic background of extreme desiccation tolerance, which is exclusively found in larvae of the only anhydrobiotic insect, Polypedilum vanderplanki. We compare the genomes of P. vanderplanki and a congeneric desiccation-sensitive midge P. nubifer. We determine that the genome of the anhydrobiotic species specifically contains clusters of multi-copy genes with products that act as molecular shields. In addition, the genome possesses several groups of genes with high similarity to known protective proteins. However, these genes are located in distinct paralogous clusters in the genome apart from the classical orthologues of the corresponding genes shared by both chironomids and other insects. The transcripts of these clustered paralogues contribute to a large majority of the mRNA pool in the desiccating larvae and most likely define successful anhydrobiosis. Comparison of expression patterns of orthologues between two chironomid species provides evidence for the existence of desiccation-specific gene expression systems in P. vanderplanki.
Figures

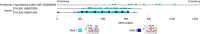
References
-
- Watanabe M. Anhydrobiosis in invertebrates. Appl. Entomol. Zool. 41, 15–31 (2006).
-
- Horikawa D. D. et al.. High hydrostatic pressure tolerance of four different anhydrobiotic animal species. Zool. Sci. 26, 238–242 (2009). - PubMed
-
- Sallon S. et al.. Germination, genetics, and growth of an ancient date seed. Science 320, 1464 (2008). - PubMed
-
- Wharton D. A. Life at the Limits: Organisms in Extreme Environments 307Cambridge Univ. Press (2002).
-
- Hinton H. E. A fly larva that tolerates dehydration and temperatures of −270° to +102°C. Nature 188, 336–337 (1960).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

