USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin
- PMID: 25216678
- PMCID: PMC4283406
- DOI: 10.15252/embj.201489729
USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin
Abstract
Mutations in the Park2 gene, encoding the E3 ubiquitin-ligase parkin, are responsible for a familial form of Parkinson's disease (PD). Parkin-mediated ubiquitination is critical for the efficient elimination of depolarized dysfunctional mitochondria by autophagy (mitophagy). As damaged mitochondria are a major source of toxic reactive oxygen species within the cell, this pathway is believed to be highly relevant to the pathogenesis of PD. Little is known about how parkin-mediated ubiquitination is regulated during mitophagy or about the nature of the ubiquitin conjugates involved. We report here that USP8/UBPY, a deubiquitinating enzyme not previously implicated in mitochondrial quality control, is critical for parkin-mediated mitophagy. USP8 preferentially removes non-canonical K6-linked ubiquitin chains from parkin, a process required for the efficient recruitment of parkin to depolarized mitochondria and for their subsequent elimination by mitophagy. This work uncovers a novel role for USP8-mediated deubiquitination of K6-linked ubiquitin conjugates from parkin in mitochondrial quality control.
Keywords: USP8; deubiquitination; mitophagy; parkin; ubiquitin.
© 2014 The Authors.
Figures
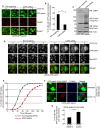
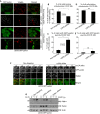

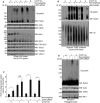

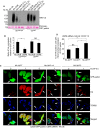
Comment in
-
DUBs counteract parkin for efficient mitophagy.EMBO J. 2014 Nov 3;33(21):2442-3. doi: 10.15252/embj.201490101. Epub 2014 Oct 1. EMBO J. 2014. PMID: 25274967 Free PMC article.
References
-
- Ali N, Zhang L, Taylor S, Mironov A, Urbe S, Woodman P. Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr Biol. 2013;23:453–461. - PubMed
-
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell. 2006;24:701–711. - PubMed
-
- de Bie P, Zaaroor-Regev D, Ciechanover A. Regulation of the Polycomb protein RING1B ubiquitination by USP7. Biochem Biophys Res Commun. 2010;400:389–395. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

