Replication fork recovery and regulation of common fragile sites stability
- PMID: 25216703
- PMCID: PMC11113654
- DOI: 10.1007/s00018-014-1718-9
Replication fork recovery and regulation of common fragile sites stability
Abstract
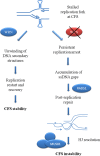
The acquisition of genomic instability is a triggering factor in cancer development, and common fragile sites (CFS) are the preferential target of chromosomal instability under conditions of replicative stress in the human genome. Although the mechanisms leading to CFS expression and the cellular factors required to suppress CFS instability remain largely undefined, it is clear that DNA becomes more susceptible to breakage when replication is impaired. The models proposed so far to explain how CFS instability arises imply that replication fork progression along these regions is perturbed due to intrinsic features of fragile sites and events that directly affect DNA replication. The observation that proteins implicated in the safe recovery of stalled forks or in engaging recombination at collapsed forks increase CFS expression when downregulated or mutated suggests that the stabilization and recovery of perturbed replication forks are crucial to guarantee CFS integrity.
Figures



References
-
- Mangelsdorf M, Ried K, Woollatt E, Dayan S, Eyre H, Finnis M, Hobson L, Nancarrow J, Venter D, Baker E, Richards RI. Chromosomal fragile site FRA16D and DNA instability in cancer. Cancer Res. 2000;60(6):1683–1689. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

