Exposing the secrets of two well-known Lactobacillus casei phages, J-1 and PL-1, by genomic and structural analysis
- PMID: 25217012
- PMCID: PMC4248993
- DOI: 10.1128/AEM.02771-14
Exposing the secrets of two well-known Lactobacillus casei phages, J-1 and PL-1, by genomic and structural analysis
Abstract
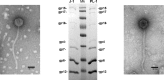
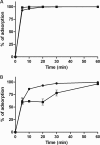
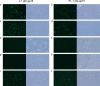
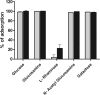
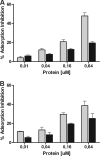
Bacteriophage J-1 was isolated in 1965 from an abnormal fermentation of Yakult using Lactobacillus casei strain Shirota, and a related phage, PL-1, was subsequently recovered from a strain resistant to J-1. Complete genome sequencing shows that J-1 and PL-1 are almost identical, but PL-1 has a deletion of 1.9 kbp relative to J-1, resulting in the loss of four predicted gene products involved in immunity regulation. The structural proteins were identified by mass spectrometry analysis. Similarly to phage A2, two capsid proteins are generated by a translational frameshift and undergo proteolytic processing. The structure of gene product 16 (gp16), a putative tail protein, was modeled based on the crystal structure of baseplate distal tail proteins (Dit) that form the baseplate hub in other Siphoviridae. However, two regions of the C terminus of gp16 could not be modeled using this template. The first region accounts for the differences between J-1 and PL-1 gp16 and showed sequence similarity to carbohydrate-binding modules (CBMs). J-1 and PL-1 GFP-gp16 fusions bind specifically to Lactobacillus casei/paracasei cells, and the addition of l-rhamnose inhibits binding. J-1 gp16 exhibited a higher affinity than PL-1 gp16 for cell walls of L. casei ATCC 27139 in phage adsorption inhibition assays, in agreement with differential adsorption kinetics observed for both phages in this strain. The data presented here provide insights into how Lactobacillus phages interact with their hosts at the first steps of infection.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Genomic Diversity of Phages Infecting Probiotic Strains of Lactobacillus paracasei.Appl Environ Microbiol. 2015 Oct 16;82(1):95-105. doi: 10.1128/AEM.02723-15. Print 2016 Jan 1. Appl Environ Microbiol. 2015. PMID: 26475105 Free PMC article.
-
Isolation and phenotypic characterization of Lactobacillus casei and Lactobacillus paracasei bacteriophage-resistant mutants.J Appl Microbiol. 2011 Aug;111(2):371-81. doi: 10.1111/j.1365-2672.2011.05056.x. Epub 2011 Jun 6. J Appl Microbiol. 2011. PMID: 21599814
-
The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria.J Bacteriol. 2002 Nov;184(21):6026-36. doi: 10.1128/JB.184.21.6026-6036.2002. J Bacteriol. 2002. PMID: 12374837 Free PMC article.
-
Bacteriophages of lactobacilli.Biochimie. 1988 Mar;70(3):401-10. doi: 10.1016/0300-9084(88)90214-3. Biochimie. 1988. PMID: 3139059 Review.
-
Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors.Viruses. 2020 May 6;12(5):512. doi: 10.3390/v12050512. Viruses. 2020. PMID: 32384698 Free PMC article. Review.
Cited by
-
Phage Adsorption to Gram-Positive Bacteria.Viruses. 2023 Jan 10;15(1):196. doi: 10.3390/v15010196. Viruses. 2023. PMID: 36680236 Free PMC article. Review.
-
The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.mBio. 2016 Jan 26;7(1):e01781-15. doi: 10.1128/mBio.01781-15. mBio. 2016. PMID: 26814179 Free PMC article.
-
Needle in a Whey-Stack: PhRACS as a Discovery Tool for Unknown Phage-Host Combinations.mBio. 2022 Feb 22;13(1):e0333421. doi: 10.1128/mbio.03334-21. Epub 2022 Jan 4. mBio. 2022. PMID: 35089052 Free PMC article.
-
Genome Sequence and Characterization of Lactobacillus casei Phage, vB_LcaM_Lbab1 Isolated from Raw Milk.Phage (New Rochelle). 2021 Mar 1;2(1):57-63. doi: 10.1089/phage.2020.0029. Epub 2021 Mar 17. Phage (New Rochelle). 2021. PMID: 36148441 Free PMC article.
-
Structure of the host-recognition device of Staphylococcus aureus phage ϕ11.Sci Rep. 2016 Jun 10;6:27581. doi: 10.1038/srep27581. Sci Rep. 2016. PMID: 27282779 Free PMC article.
References
-
- Suarez VB, Quiberoni A, Binetti AG, Reinheimer JA. 2002. Thermophilic lactic acid bacteria phages isolated from Argentinian dairy industries. J. Food Prot. 65:1597–1604. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

