The cytoplasmic poly(A) polymerases GLD-2 and GLD-4 promote general gene expression via distinct mechanisms
- PMID: 25217583
- PMCID: PMC4191412
- DOI: 10.1093/nar/gku838
The cytoplasmic poly(A) polymerases GLD-2 and GLD-4 promote general gene expression via distinct mechanisms
Abstract
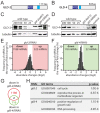
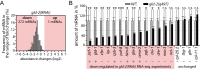
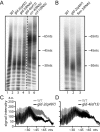
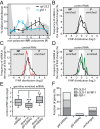
Post-transcriptional gene regulation mechanisms decide on cellular mRNA activities. Essential gatekeepers of post-transcriptional mRNA regulation are broadly conserved mRNA-modifying enzymes, such as cytoplasmic poly(A) polymerases (cytoPAPs). Although these non-canonical nucleotidyltransferases efficiently elongate mRNA poly(A) tails in artificial tethering assays, we still know little about their global impact on poly(A) metabolism and their individual molecular roles in promoting protein production in organisms. Here, we use the animal model Caenorhabditis elegans to investigate the global mechanisms of two germline-enriched cytoPAPs, GLD-2 and GLD-4, by combining polysome profiling with RNA sequencing. Our analyses suggest that GLD-2 activity mediates mRNA stability of many translationally repressed mRNAs. This correlates with a general shortening of long poly(A) tails in gld-2-compromised animals, suggesting that most if not all targets are stabilized via robust GLD-2-mediated polyadenylation. By contrast, only mild polyadenylation defects are found in gld-4-compromised animals and few mRNAs change in abundance. Interestingly, we detect a reduced number of polysomes in gld-4 mutants and GLD-4 protein co-sediments with polysomes, which together suggest that GLD-4 might stimulate or maintain translation directly. Our combined data show that distinct cytoPAPs employ different RNA-regulatory mechanisms to promote gene expression, offering new insights into translational activation of mRNAs.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Polyadenylation is the key aspect of GLD-2 function in C. elegans.RNA. 2017 Aug;23(8):1180-1187. doi: 10.1261/rna.061473.117. Epub 2017 May 10. RNA. 2017. PMID: 28490506 Free PMC article.
-
GLD-4-mediated translational activation regulates the size of the proliferative germ cell pool in the adult C. elegans germ line.PLoS Genet. 2014 Sep 25;10(9):e1004647. doi: 10.1371/journal.pgen.1004647. eCollection 2014 Sep. PLoS Genet. 2014. PMID: 25254367 Free PMC article.
-
The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line.Proc Natl Acad Sci U S A. 2006 Oct 10;103(41):15108-12. doi: 10.1073/pnas.0607050103. Epub 2006 Sep 29. Proc Natl Acad Sci U S A. 2006. PMID: 17012378 Free PMC article.
-
Non-canonical poly(A) polymerase in mammalian gametogenesis.Biochim Biophys Acta. 2008 Apr;1779(4):230-8. doi: 10.1016/j.bbagrm.2008.01.004. Epub 2008 Feb 12. Biochim Biophys Acta. 2008. PMID: 18294465 Review.
-
[Regulation of spermatogenesis by polyadenylation and deadenylation of mRNAs].Tanpakushitsu Kakusan Koso. 2009 Dec;54(16 Suppl):2073-9. Tanpakushitsu Kakusan Koso. 2009. PMID: 21089620 Review. Japanese. No abstract available.
Cited by
-
The endogenous mex-3 3´UTR is required for germline repression and contributes to optimal fecundity in C. elegans.PLoS Genet. 2021 Aug 23;17(8):e1009775. doi: 10.1371/journal.pgen.1009775. eCollection 2021 Aug. PLoS Genet. 2021. PMID: 34424904 Free PMC article.
-
Polyadenylation is the key aspect of GLD-2 function in C. elegans.RNA. 2017 Aug;23(8):1180-1187. doi: 10.1261/rna.061473.117. Epub 2017 May 10. RNA. 2017. PMID: 28490506 Free PMC article.
-
Mechanisms of microRNA turnover.Curr Opin Plant Biol. 2015 Oct;27:199-206. doi: 10.1016/j.pbi.2015.07.008. Epub 2015 Sep 2. Curr Opin Plant Biol. 2015. PMID: 26342825 Free PMC article. Review.
-
Germline immortality relies on TRIM32-mediated turnover of a maternal mRNA activator in C. elegans.Sci Adv. 2022 Oct 14;8(41):eabn0897. doi: 10.1126/sciadv.abn0897. Epub 2022 Oct 14. Sci Adv. 2022. PMID: 36240265 Free PMC article.
-
Biology of the Caenorhabditis elegans Germline Stem Cell System.Genetics. 2019 Dec;213(4):1145-1188. doi: 10.1534/genetics.119.300238. Genetics. 2019. PMID: 31796552 Free PMC article. Review.
References
-
- Sachs A., Wahle E. Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem. 1993;268:22955–22958. - PubMed
-
- Doma M.K., Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. - PubMed
-
- Salles F.J., Lieberfarb M.E., Wreden C., Gergen J.P., Strickland S. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science. 1994;266:1996–1999. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

