Ms1, a novel sRNA interacting with the RNA polymerase core in mycobacteria
- PMID: 25217589
- PMCID: PMC4191392
- DOI: 10.1093/nar/gku793
Ms1, a novel sRNA interacting with the RNA polymerase core in mycobacteria
Abstract
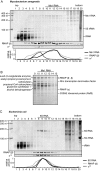
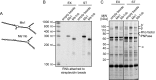
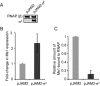
Small RNAs (sRNAs) are molecules essential for a number of regulatory processes in the bacterial cell. Here we characterize Ms1, a sRNA that is highly expressed in Mycobacterium smegmatis during stationary phase of growth. By glycerol gradient ultracentrifugation, RNA binding assay, and RNA co-immunoprecipitation, we show that Ms1 interacts with the RNA polymerase (RNAP) core that is free of the primary sigma factor (σA) or any other σ factor. This contrasts with the situation in most other species where it is 6S RNA that interacts with RNAP and this interaction requires the presence of σA. The difference in the interaction of the two types of sRNAs (Ms1 or 6S RNA) with RNAP possibly reflects the difference in the composition of the transcriptional machinery between mycobacteria and other species. Unlike Escherichia coli, stationary phase M. smegmatis cells contain relatively few RNAP molecules in complex with σA. Thus, Ms1 represents a novel type of small RNAs interacting with RNAP.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


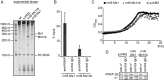
Similar articles
-
Ms1 RNA increases the amount of RNA polymerase in Mycobacterium smegmatis.Mol Microbiol. 2019 Feb;111(2):354-372. doi: 10.1111/mmi.14159. Epub 2018 Dec 11. Mol Microbiol. 2019. PMID: 30427073
-
RIP-seq reveals RNAs that interact with RNA polymerase and primary sigma factors in bacteria.Nucleic Acids Res. 2024 May 8;52(8):4604-4626. doi: 10.1093/nar/gkae081. Nucleic Acids Res. 2024. PMID: 38348908 Free PMC article.
-
MoaB2, a newly identified transcription factor, binds to σA in Mycobacterium smegmatis.J Bacteriol. 2024 Dec 19;206(12):e0006624. doi: 10.1128/jb.00066-24. Epub 2024 Nov 5. J Bacteriol. 2024. PMID: 39499088 Free PMC article.
-
Small Noncoding 6S RNAs of Bacteria.Biochemistry (Mosc). 2015 Nov;80(11):1429-46. doi: 10.1134/S0006297915110048. Biochemistry (Mosc). 2015. PMID: 26615434 Review.
-
6S RNA, a Global Regulator of Transcription.Microbiol Spectr. 2018 May;6(3):10.1128/microbiolspec.rwr-0019-2018. doi: 10.1128/microbiolspec.RWR-0019-2018. Microbiol Spectr. 2018. PMID: 29916345 Free PMC article. Review.
Cited by
-
Ms1 RNA Interacts With the RNA Polymerase Core in Streptomyces coelicolor and Was Identified in Majority of Actinobacteria Using a Linguistic Gene Synteny Search.Front Microbiol. 2022 May 11;13:848536. doi: 10.3389/fmicb.2022.848536. eCollection 2022. Front Microbiol. 2022. PMID: 35633709 Free PMC article.
-
Conservation of Small Regulatory RNAs in Vibrio parahaemolyticus: Possible role of RNA-OUT Encoded by the Pathogenicity Island (VPaI-7) of Pandemic Strains.Int J Mol Sci. 2019 Jun 10;20(11):2827. doi: 10.3390/ijms20112827. Int J Mol Sci. 2019. PMID: 31185635 Free PMC article.
-
Comparative genomics of Mycobacterium mucogenicum and Mycobacterium neoaurum clade members emphasizing tRNA and non-coding RNA.BMC Evol Biol. 2019 Jun 18;19(1):124. doi: 10.1186/s12862-019-1447-7. BMC Evol Biol. 2019. PMID: 31215393 Free PMC article.
-
RNA atlas of human bacterial pathogens uncovers stress dynamics linked to infection.Nat Commun. 2021 Jun 2;12(1):3282. doi: 10.1038/s41467-021-23588-w. Nat Commun. 2021. PMID: 34078900 Free PMC article.
-
Small RNAs Asserting Big Roles in Mycobacteria.Noncoding RNA. 2021 Oct 29;7(4):69. doi: 10.3390/ncrna7040069. Noncoding RNA. 2021. PMID: 34842799 Free PMC article. Review.
References
-
- Stewart G.R., Robertson B.D., Young D.B. Tuberculosis: a problem with persistence. Nat. Rev. Microbiol. 2003;1:97–105. - PubMed
-
- Waagmeester A., Thompson J., Reyrat J.M. Identifying sigma factors in Mycobacterium smegmatis by comparative genomic analysis. Trends Microbiol. 2005;13:505–509. - PubMed
-
- Sharma U.K., Chatterji D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol. Rev. 2010;34:646–657. - PubMed
-
- Österberg S., del Peso-Santos T., Shingler V. Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 2011;65:37–55. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

