Topological constraints are major determinants of tRNA tertiary structure and dynamics and provide basis for tertiary folding cooperativity
- PMID: 25217593
- PMCID: PMC4191394
- DOI: 10.1093/nar/gku807
Topological constraints are major determinants of tRNA tertiary structure and dynamics and provide basis for tertiary folding cooperativity
Abstract
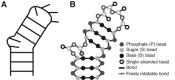
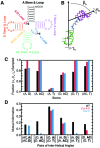
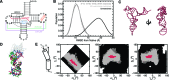
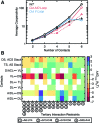
Recent studies have shown that basic steric and connectivity constraints encoded at the secondary structure level are key determinants of 3D structure and dynamics in simple two-way RNA junctions. However, the role of these topological constraints in higher order RNA junctions remains poorly understood. Here, we use a specialized coarse-grained molecular dynamics model to directly probe the thermodynamic contributions of topological constraints in defining the 3D architecture and dynamics of transfer RNA (tRNA). Topological constraints alone restrict tRNA's allowed conformational space by over an order of magnitude and strongly discriminate against formation of non-native tertiary contacts, providing a sequence independent source of folding specificity. Topological constraints also give rise to long-range correlations between the relative orientation of tRNA's helices, which in turn provides a mechanism for encoding thermodynamic cooperativity between distinct tertiary interactions. These aspects of topological constraints make it such that only several tertiary interactions are needed to confine tRNA to its native global structure and specify functionally important 3D dynamics. We further show that topological constraints are conserved across tRNA's different naturally occurring secondary structures. Taken together, our results emphasize the central role of secondary-structure-encoded topological constraints in defining RNA 3D structure, dynamics and folding.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Noncanonical secondary structure stabilizes mitochondrial tRNA(Ser(UCN)) by reducing the entropic cost of tertiary folding.J Am Chem Soc. 2015 Mar 18;137(10):3592-9. doi: 10.1021/ja5130308. Epub 2015 Mar 9. J Am Chem Soc. 2015. PMID: 25705930 Free PMC article.
-
Secondary structure encodes a cooperative tertiary folding funnel in the Azoarcus ribozyme.Nucleic Acids Res. 2016 Jan 8;44(1):402-12. doi: 10.1093/nar/gkv1055. Epub 2015 Oct 19. Nucleic Acids Res. 2016. PMID: 26481360 Free PMC article.
-
Coarse grained models reveal essential contributions of topological constraints to the conformational free energy of RNA bulges.J Phys Chem B. 2014 Mar 13;118(10):2615-27. doi: 10.1021/jp411478x. Epub 2014 Mar 3. J Phys Chem B. 2014. PMID: 24547945 Free PMC article.
-
Topological constraints: using RNA secondary structure to model 3D conformation, folding pathways, and dynamic adaptation.Curr Opin Struct Biol. 2011 Jun;21(3):296-305. doi: 10.1016/j.sbi.2011.03.009. Epub 2011 Apr 14. Curr Opin Struct Biol. 2011. PMID: 21497083 Free PMC article. Review.
-
MD Simulations of tRNA and Aminoacyl-tRNA Synthetases: Dynamics, Folding, Binding, and Allostery.Int J Mol Sci. 2015 Jul 13;16(7):15872-902. doi: 10.3390/ijms160715872. Int J Mol Sci. 2015. PMID: 26184179 Free PMC article. Review.
Cited by
-
Capturing RNA Folding Free Energy with Coarse-Grained Molecular Dynamics Simulations.Sci Rep. 2017 Apr 10;7:45812. doi: 10.1038/srep45812. Sci Rep. 2017. PMID: 28393861 Free PMC article.
-
Tuning RNA folding and function through rational design of junction topology.Nucleic Acids Res. 2017 Sep 19;45(16):9706-9715. doi: 10.1093/nar/gkx614. Nucleic Acids Res. 2017. PMID: 28934478 Free PMC article.
-
Principles and Overview of Sampling Methods for Modeling Macromolecular Structure and Dynamics.PLoS Comput Biol. 2016 Apr 28;12(4):e1004619. doi: 10.1371/journal.pcbi.1004619. eCollection 2016 Apr. PLoS Comput Biol. 2016. PMID: 27124275 Free PMC article. Review.
-
Noncanonical secondary structure stabilizes mitochondrial tRNA(Ser(UCN)) by reducing the entropic cost of tertiary folding.J Am Chem Soc. 2015 Mar 18;137(10):3592-9. doi: 10.1021/ja5130308. Epub 2015 Mar 9. J Am Chem Soc. 2015. PMID: 25705930 Free PMC article.
-
Hierarchy of RNA functional dynamics.Annu Rev Biochem. 2014;83:441-66. doi: 10.1146/annurev-biochem-060713-035524. Epub 2014 Mar 5. Annu Rev Biochem. 2014. PMID: 24606137 Free PMC article. Review.
References
-
- Gesteland R.F., Cech T.R., Atkins J.F. The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA World. Plainview, NY: Cold Spring Harbor Laboratory Press; 2006.
-
- Cruz J.A., Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. - PubMed
-
- Thirumalai D., Hyeon C. Theory of RNA folding: from hairpins to ribozymes. In: Walter NG, Woodson SA, Batey R, editors. Non-Protein Coding RNAs. Vol. 13. Berlin, Germany: Springer-Verlag; 2009. pp. 27–47.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

