Serine integrase chimeras with activity in E. coli and HeLa cells
- PMID: 25217617
- PMCID: PMC4197438
- DOI: 10.1242/bio.20148748
Serine integrase chimeras with activity in E. coli and HeLa cells
Abstract
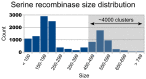
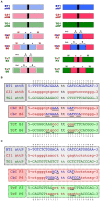
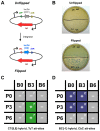
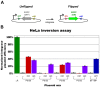
In recent years, application of serine integrases for genomic engineering has increased in popularity. The factor-independence and unidirectionality of these large serine recombinases makes them well suited for reactions such as site-directed vector integration and cassette exchange in a wide variety of organisms. In order to generate information that might be useful for altering the specificity of serine integrases and to improve their efficiency, we tested a hybridization strategy that has been successful with several small serine recombinases. We created chimeras derived from three characterized members of the serine integrase family, phiC31, phiBT1, and TG1 integrases, by joining their amino- and carboxy-terminal portions. We found that several phiBT1-phiC31 (BC) and phiC31-TG1 (CT) hybrid integrases are active in E. coli. BC chimeras function on native att-sites and on att-sites that are hybrids between those of the two donor enzymes, while CT chimeras only act on the latter att-sites. A BC hybrid, BC{-1}, was also active in human HeLa cells. Our work is the first to demonstrate chimeric serine integrase activity. This analysis sheds light on integrase structure and function, and establishes a potentially tractable means to probe the specificity of the thousands of putative large serine recombinases that have been revealed by bioinformatics studies.
Keywords: Chimeric recombinase; Genome engineering; Hybrid protein; Phage integrase; Sequence-specific recombination.
© 2014. Published by The Company of Biologists Ltd.
Conflict of interest statement
Figures



Similar articles
-
The site-specific integration reaction of Listeria phage A118 integrase, a serine recombinase.Mob DNA. 2013 Jan 3;4(1):2. doi: 10.1186/1759-8753-4-2. Mob DNA. 2013. PMID: 23282060 Free PMC article.
-
Using phage integrases in a site-specific dual integrase cassette exchange strategy.Methods Mol Biol. 2015;1239:29-38. doi: 10.1007/978-1-4939-1862-1_3. Methods Mol Biol. 2015. PMID: 25408400
-
The ΦBT1 large serine recombinase catalyzes DNA integration at pseudo-attB sites in the genus Nocardia.PeerJ. 2018 May 4;6:e4784. doi: 10.7717/peerj.4784. eCollection 2018. PeerJ. 2018. PMID: 29740520 Free PMC article.
-
Phage integrases: biology and applications.J Mol Biol. 2004 Jan 16;335(3):667-78. doi: 10.1016/j.jmb.2003.09.082. J Mol Biol. 2004. PMID: 14687564 Review.
-
Phage-encoded Serine Integrases and Other Large Serine Recombinases.Microbiol Spectr. 2015 Aug;3(4). doi: 10.1128/microbiolspec.MDNA3-0059-2014. Microbiol Spectr. 2015. PMID: 26350324 Review.
Cited by
-
Mechanism of bacterial gene rearrangement: SprA-catalyzed precise DNA recombination and its directionality control by SprB ensure the gene rearrangement and stable expression of spsM during sporulation in Bacillus subtilis.Nucleic Acids Res. 2017 Jun 20;45(11):6669-6683. doi: 10.1093/nar/gkx466. Nucleic Acids Res. 2017. PMID: 28535266 Free PMC article.
-
Diverse target gene modifications in Plasmodium falciparum using Bxb1 integrase and an intronic attB.Parasit Vectors. 2018 Oct 17;11(1):548. doi: 10.1186/s13071-018-3129-5. Parasit Vectors. 2018. PMID: 30333047 Free PMC article. Review.
-
Control of serine integrase recombination directionality by fusion with the directionality factor.Nucleic Acids Res. 2017 Aug 21;45(14):8635-8645. doi: 10.1093/nar/gkx567. Nucleic Acids Res. 2017. PMID: 28666339 Free PMC article.
-
A novel family of tyrosine integrases encoded by the temperate pleolipovirus SNJ2.Nucleic Acids Res. 2018 Mar 16;46(5):2521-2536. doi: 10.1093/nar/gky005. Nucleic Acids Res. 2018. PMID: 29361162 Free PMC article.
-
Systematic discovery of recombinases for efficient integration of large DNA sequences into the human genome.Nat Biotechnol. 2023 Apr;41(4):488-499. doi: 10.1038/s41587-022-01494-w. Epub 2022 Oct 10. Nat Biotechnol. 2023. PMID: 36217031 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

