Autocrine Activation of the Wnt/β-Catenin Pathway by CUX1 and GLIS1 in Breast Cancers
- PMID: 25217618
- PMCID: PMC4197442
- DOI: 10.1242/bio.20148193
Autocrine Activation of the Wnt/β-Catenin Pathway by CUX1 and GLIS1 in Breast Cancers
Abstract
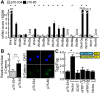
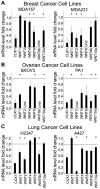
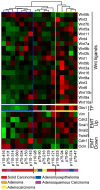
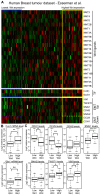
Autocrine activation of the Wnt/β-catenin pathway occurs in several cancers, notably in breast tumors, and is associated with higher expression of various Wnt ligands. Using various inhibitors of the FZD/LRP receptor complex, we demonstrate that some adenosquamous carcinomas that develop in MMTV-CUX1 transgenic mice represent a model for autocrine activation of the Wnt/β-catenin pathway. By comparing expression profiles of laser-capture microdissected mammary tumors, we identify Glis1 as a transcription factor that is highly expressed in the subset of tumors with elevated Wnt gene expression. Analysis of human cancer datasets confirms that elevated WNT gene expression is associated with high levels of CUX1 and GLIS1 and correlates with genes of the epithelial-to-mesenchymal transition (EMT) signature: VIM, SNAI1 and TWIST1 are elevated whereas CDH1 and OCLN are decreased. Co-expression experiments demonstrate that CUX1 and GLIS1 cooperate to stimulate TCF/β-catenin transcriptional activity and to enhance cell migration and invasion. Altogether, these results provide additional evidence for the role of GLIS1 in reprogramming gene expression and suggest a hierarchical model for transcriptional regulation of the Wnt/β-catenin pathway and the epithelial-to-mesenchymal transition.
Keywords: Breast cancer; Expression profiling; Mouse tumor model; Transcriptional regulation; Wnt/β-Catenin pathway.
© 2014. Published by The Company of Biologists Ltd.
Conflict of interest statement
Figures



Similar articles
-
Mouse mammary tumor virus p75 and p110 CUX1 transgenic mice develop mammary tumors of various histologic types.Cancer Res. 2009 Sep 15;69(18):7188-97. doi: 10.1158/0008-5472.CAN-08-4899. Epub 2009 Sep 8. Cancer Res. 2009. PMID: 19738070
-
GPNMB augments Wnt-1 mediated breast tumor initiation and growth by enhancing PI3K/AKT/mTOR pathway signaling and β-catenin activity.Oncogene. 2019 Jun;38(26):5294-5307. doi: 10.1038/s41388-019-0793-7. Epub 2019 Mar 26. Oncogene. 2019. PMID: 30914799
-
Activation of the canonical Wnt/β-catenin pathway in ATF3-induced mammary tumors.PLoS One. 2011 Jan 31;6(1):e16515. doi: 10.1371/journal.pone.0016515. PLoS One. 2011. PMID: 21304988 Free PMC article.
-
Cross-talk of WNT and FGF signaling pathways at GSK3beta to regulate beta-catenin and SNAIL signaling cascades.Cancer Biol Ther. 2006 Sep;5(9):1059-64. doi: 10.4161/cbt.5.9.3151. Epub 2006 Sep 4. Cancer Biol Ther. 2006. PMID: 16940750 Review.
-
A Wnt-ow of opportunity: targeting the Wnt/beta-catenin pathway in breast cancer.Curr Drug Targets. 2010 Sep;11(9):1074-88. doi: 10.2174/138945010792006780. Curr Drug Targets. 2010. PMID: 20545611 Review.
Cited by
-
GLIS1-3: Links to Primary Cilium, Reprogramming, Stem Cell Renewal, and Disease.Cells. 2022 Jun 3;11(11):1833. doi: 10.3390/cells11111833. Cells. 2022. PMID: 35681527 Free PMC article. Review.
-
GLIS Family Zinc Finger 1 was First Linked With Preaxial Polydactyly I in Humans by Stepwise Genetic Analysis.Front Cell Dev Biol. 2022 Jan 11;9:781388. doi: 10.3389/fcell.2021.781388. eCollection 2021. Front Cell Dev Biol. 2022. PMID: 35087831 Free PMC article.
-
Lack of APC somatic mutation is associated with early-onset colorectal cancer in African Americans.Carcinogenesis. 2018 Dec 13;39(11):1331-1341. doi: 10.1093/carcin/bgy122. Carcinogenesis. 2018. PMID: 30239619 Free PMC article.
-
GLIS1 in Cancer-Associated Fibroblasts Regulates the Migration and Invasion of Ovarian Cancer Cells.Int J Mol Sci. 2022 Feb 17;23(4):2218. doi: 10.3390/ijms23042218. Int J Mol Sci. 2022. PMID: 35216340 Free PMC article.
-
BAG3 Overexpression and Cytoprotective Autophagy Mediate Apoptosis Resistance in Chemoresistant Breast Cancer Cells.Neoplasia. 2018 Mar;20(3):263-279. doi: 10.1016/j.neo.2018.01.001. Epub 2018 Feb 22. Neoplasia. 2018. PMID: 29462756 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

