Uncovering global SUMOylation signaling networks in a site-specific manner
- PMID: 25218447
- PMCID: PMC4259010
- DOI: 10.1038/nsmb.2890
Uncovering global SUMOylation signaling networks in a site-specific manner
Abstract
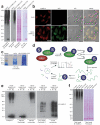
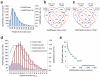
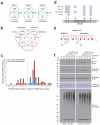
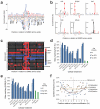
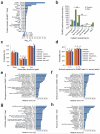
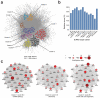
SUMOylation is a reversible post-translational modification essential for genome stability. Using high-resolution MS, we have studied global SUMOylation in human cells in a site-specific manner, identifying a total of >4,300 SUMOylation sites in >1,600 proteins. To our knowledge, this is the first time that >1,000 SUMOylation sites have been identified under standard growth conditions. We quantitatively studied SUMOylation dynamics in response to SUMO protease inhibition, proteasome inhibition and heat shock. Many SUMOylated lysines have previously been reported to be ubiquitinated, acetylated or methylated, thus indicating cross-talk between SUMO and other post-translational modifications. We identified 70 phosphorylation and four acetylation events in proximity to SUMOylation sites, and we provide evidence for acetylation-dependent SUMOylation of endogenous histone H3. SUMOylation regulates target proteins involved in all nuclear processes including transcription, DNA repair, chromatin remodeling, precursor-mRNA splicing and ribosome assembly.
Figures
References
Reference List
-
- Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 2013;49:795–807. - PubMed
-
- Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 2010;11:479–489. - PubMed
-
- Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005;15:536–541. - PubMed
-
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. - PubMed
-
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. - PubMed
References for Online Methods
-
- Vellinga J, et al. A system for efficient generation of adenovirus protein IX-producing helper cell lines. J. Gene Med. 2006;8:147–154. - PubMed
-
- Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat. Protoc. 2006;1:241–245. - PubMed
-
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. - PubMed
-
- Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome. Res. 2011;10:1794–1805. - PubMed
-
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

