Preanalytical variables and performance of diagnostic RNA-based gene expression analysis in breast cancer
- PMID: 25218890
- PMCID: PMC4180906
- DOI: 10.1007/s00428-014-1652-0
Preanalytical variables and performance of diagnostic RNA-based gene expression analysis in breast cancer
Abstract
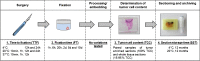
Prognostic multigene expression assays have become widely available to provide additional information to standard clinical parameters and to support clinicians in treatment decisions. In this study, we analyzed the impact of variations in tissue handling on the diagnostic EndoPredict test results. EndoPredict is a quantitative reverse transcription PCR assay conducted on RNA from formalin-fixed, paraffin-embedded (FFPE) tissue that predicts the likelihood of distant recurrence in patients with ER-positive/HER2-negative breast cancer. In this study, we performed a total of 138 EndoPredict assays to study the effects of preanalytical variables such as time to fixation, fixation time, tumor cell content, and section storage time on the EndoPredict test results. A time to fixation of up to 12 h and fixation of up to 5 days did not affect the results of the gene expression test. Paired samples of FFPE sections with tumor cell content ranging from 15 to 95 % and tumor-enriched samples showed a correlation coefficient of 0.97. Test results of tissue sections that have been stored for 12 months at +4 or +20 °C showed a correlation of 0.99 when compared to results of nonstored sections. In conclusion, preanalytical tissue handling is not a critical factor for diagnostic gene expression analysis with the EndoPredict assay. The test can therefore be easily integrated into the standard workflow of molecular pathology.
Figures




Similar articles
-
Decentral gene expression analysis for ER+/Her2- breast cancer: results of a proficiency testing program for the EndoPredict assay.Virchows Arch. 2012 Mar;460(3):251-9. doi: 10.1007/s00428-012-1204-4. Epub 2012 Feb 28. Virchows Arch. 2012. PMID: 22371223 Free PMC article.
-
Decentral gene expression analysis: analytical validation of the Endopredict genomic multianalyte breast cancer prognosis test.BMC Cancer. 2012 Oct 5;12:456. doi: 10.1186/1471-2407-12-456. BMC Cancer. 2012. PMID: 23039280 Free PMC article.
-
Comparison of gene expression profiling by reverse transcription quantitative PCR between fresh frozen and formalin-fixed, paraffin-embedded breast cancer tissues.Biotechniques. 2010 May;48(5):389-97. doi: 10.2144/000113388. Biotechniques. 2010. PMID: 20569212
-
A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: how well do you know your FFPE specimen?Arch Pathol Lab Med. 2014 Nov;138(11):1520-30. doi: 10.5858/arpa.2013-0691-RA. Arch Pathol Lab Med. 2014. PMID: 25357115 Review.
-
Tissue handling and specimen preparation in surgical pathology: issues concerning the recovery of nucleic acids from formalin-fixed, paraffin-embedded tissue.Arch Pathol Lab Med. 2008 Dec;132(12):1929-35. doi: 10.5858/132.12.1929. Arch Pathol Lab Med. 2008. PMID: 19061293 Review.
Cited by
-
Impact of different stabilization methods on RT-qPCR results using human lung tissue samples.Sci Rep. 2020 Feb 27;10(1):3579. doi: 10.1038/s41598-020-60618-x. Sci Rep. 2020. PMID: 32108147 Free PMC article.
-
Small Nucleolar RNA Score: An Assay to Detect Formalin-Overfixed Tissue.Biopreserv Biobank. 2018 Dec;16(6):467-476. doi: 10.1089/bio.2018.0042. Epub 2018 Sep 19. Biopreserv Biobank. 2018. PMID: 30234371 Free PMC article.
-
[Molecular pathology for breast cancer: Importance of the gene expression profile].Pathologe. 2015 Mar;36(2):145-53. doi: 10.1007/s00292-015-0009-z. Pathologe. 2015. PMID: 25836324 Review. German.
-
Robustness of biomarker determination in breast cancer by RT-qPCR: impact of tumor cell content, DCIS and non-neoplastic breast tissue.Diagn Pathol. 2018 Oct 20;13(1):83. doi: 10.1186/s13000-018-0760-6. Diagn Pathol. 2018. PMID: 30342538 Free PMC article.
-
Quality Considerations When Using Tissue Samples for Biomarker Studies in Cancer Research.Biomark Insights. 2021 Apr 20;16:11772719211009513. doi: 10.1177/11772719211009513. eCollection 2021. Biomark Insights. 2021. PMID: 33958852 Free PMC article.
References
-
- Filipits M, Rudas M, Jakesz R, Dubsky P, Fitzal F, Singer CF, Dietze O, Greil R, Jelen A, Sevelda P, Freibauer C, Muller V, Janicke F, Schmidt M, Kolbl H, Rody A, Kaufmann M, Schroth W, Brauch H, Schwab M, Fritz P, Weber KE, Feder IS, Hennig G, Kronenwett R, Gehrmann M, Gnant M. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors Clin. Cancer Res. 2011;17:6012–6020. - PubMed
-
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer N. Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. - DOI - PubMed
-
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes J. Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. - DOI - PMC - PubMed
-
- van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

