BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork
- PMID: 25219499
- PMCID: PMC4185004
- DOI: 10.1016/j.molcel.2014.08.012
BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork
Abstract
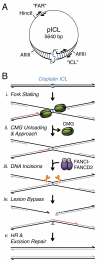
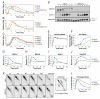
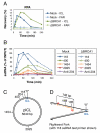
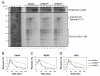
The tumor suppressor protein BRCA1 promotes homologous recombination (HR), a high-fidelity mechanism to repair DNA double-strand breaks (DSBs) that arise during normal replication and in response to DNA-damaging agents. Recent genetic experiments indicate that BRCA1 also performs an HR-independent function during the repair of DNA interstrand crosslinks (ICLs). Here we show that BRCA1 is required to unload the CMG helicase complex from chromatin after replication forks collide with an ICL. Eviction of the stalled helicase allows leading strands to be extended toward the ICL, followed by endonucleolytic processing of the crosslink, lesion bypass, and DSB repair. Our results identify BRCA1-dependent helicase unloading as a critical, early event in ICL repair.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Replication Fork Reversal during DNA Interstrand Crosslink Repair Requires CMG Unloading.Cell Rep. 2018 Jun 19;23(12):3419-3428. doi: 10.1016/j.celrep.2018.05.061. Cell Rep. 2018. PMID: 29924986 Free PMC article.
-
p97 Promotes a Conserved Mechanism of Helicase Unloading during DNA Cross-Link Repair.Mol Cell Biol. 2016 Nov 14;36(23):2983-2994. doi: 10.1128/MCB.00434-16. Print 2016 Dec 1. Mol Cell Biol. 2016. PMID: 27644328 Free PMC article.
-
Single-strand DNA breaks cause replisome disassembly.Mol Cell. 2021 Mar 18;81(6):1309-1318.e6. doi: 10.1016/j.molcel.2020.12.039. Epub 2021 Jan 22. Mol Cell. 2021. PMID: 33484638 Free PMC article.
-
Revisiting the BRCA-pathway through the lens of replication gap suppression: "Gaps determine therapy response in BRCA mutant cancer".DNA Repair (Amst). 2021 Nov;107:103209. doi: 10.1016/j.dnarep.2021.103209. Epub 2021 Aug 13. DNA Repair (Amst). 2021. PMID: 34419699 Free PMC article. Review.
-
Moonlighting at replication forks - a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51.FEBS Lett. 2017 Apr;591(8):1083-1100. doi: 10.1002/1873-3468.12556. Epub 2017 Jan 30. FEBS Lett. 2017. PMID: 28079255 Review.
Cited by
-
DNA replication: Mechanisms and therapeutic interventions for diseases.MedComm (2020). 2023 Feb 5;4(1):e210. doi: 10.1002/mco2.210. eCollection 2023 Feb. MedComm (2020). 2023. PMID: 36776764 Free PMC article. Review.
-
BRCA1 and NORE1A Form a Her2/Ras Regulated Tumor Suppressor Complex Modulating Senescence.Cancers (Basel). 2023 Aug 16;15(16):4133. doi: 10.3390/cancers15164133. Cancers (Basel). 2023. PMID: 37627161 Free PMC article.
-
MicroRNA-34a promotes genomic instability by a broad suppression of genome maintenance mechanisms downstream of the oncogene KSHV-vGPCR.Oncotarget. 2016 Mar 1;7(9):10414-32. doi: 10.18632/oncotarget.7248. Oncotarget. 2016. PMID: 26871287 Free PMC article.
-
Mechanistic insights into how CMG helicase facilitates replication past DNA roadblocks.DNA Repair (Amst). 2017 Jul;55:76-82. doi: 10.1016/j.dnarep.2017.05.005. Epub 2017 May 20. DNA Repair (Amst). 2017. PMID: 28554039 Free PMC article. Review.
-
The evolving role of DNA inter-strand crosslinks in chemotherapy.Curr Opin Pharmacol. 2018 Aug;41:20-26. doi: 10.1016/j.coph.2018.04.004. Epub 2018 Apr 18. Curr Opin Pharmacol. 2018. PMID: 29679802 Free PMC article. Review.
References
-
- Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair. 2010;9:1219–1228. - PubMed
-
- Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The Breast Cancer Susceptibility Gene BRCA1 Is Required for Subnuclear Assembly of Rad51 and Survival following Treatment with the DNA Cross-linking Agent Cisplatin. Journal of Biological Chemistry. 2000;275:23899–23903. - PubMed
-
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature Structural & Molecular Biology. 2010;17:688–695. - PMC - PubMed
-
- Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nature Genetics. 2005;37:953–957. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

