Ral small GTPase signaling and oncogenesis: More than just 15minutes of fame
- PMID: 25219551
- PMCID: PMC4201770
- DOI: 10.1016/j.bbamcr.2014.09.004
Ral small GTPase signaling and oncogenesis: More than just 15minutes of fame
Abstract
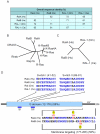
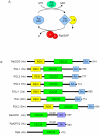
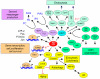
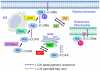
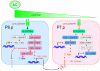
Since their discovery in 1986, Ral (Ras-like) GTPases have emerged as critical regulators of diverse cellular functions. Ral-selective guanine nucleotide exchange factors (RalGEFs) function as downstream effectors of the Ras oncoprotein, and the RalGEF-Ral signaling network comprises the third best characterized effector of Ras-dependent human oncogenesis. Because of this, Ral GTPases as well as their effectors are being explored as possible therapeutic targets in the treatment of RAS mutant cancer. The two Ral isoforms, RalA and RalB, interact with a variety of downstream effectors and have been found to play key and distinct roles in both normal and neoplastic cell physiology including regulation of vesicular trafficking, migration and invasion, tumor formation, metastasis, and gene expression. In this review we provide an overview of Ral biochemistry and biology, and we highlight recent discoveries.
Keywords: Exocyst; GTPase activating protein; Guanine nucleotide exchange factor; Ras; Rho; Small GTPase.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures

References
-
- Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

