Identification and characterization of new family members in the tautomerase superfamily: analysis and implications
- PMID: 25219626
- PMCID: PMC4258425
- DOI: 10.1016/j.abb.2014.08.019
Identification and characterization of new family members in the tautomerase superfamily: analysis and implications
Abstract
Tautomerase superfamily members are characterized by a β-α-β building block and a catalytic amino terminal proline. 4-Oxalocrotonate tautomerase (4-OT) and malonate semialdehyde decarboxylase (MSAD) are the title enzymes of two of the five known families in the superfamily. Two recent developments in these families indicate that there might be more metabolic diversity in the tautomerase superfamily than previously thought. 4-OT homologues have been identified in three biosynthetic pathways, whereas all previously characterized 4-OTs are found in catabolic pathways. In the MSAD family, homologues have been characterized that lack decarboxylase activity, but have a modest hydratase activity using 2-oxo-3-pentynoate. This observation stands in contrast to the first characterized MSAD, which is a proficient decarboxylase and a less efficient hydratase. The hydratase activity was thought to be a vestigial and promiscuous activity. However, this recent discovery suggests that the hydratase activity might reflect a new activity in the MSAD family for an unknown substrate. These discoveries open up new avenues of research in the tautomerase superfamily.
Keywords: 4-Oxalocrotonate tautomerase; Catalytic amino terminal proline; Catalytic promiscuity; Hydratase activity; Malonate semialdehyde decarboxylase; Tautomerase superfamily; β–α–β fold.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
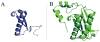






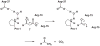
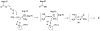
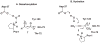
Similar articles
-
The chemical versatility of the beta-alpha-beta fold: catalytic promiscuity and divergent evolution in the tautomerase superfamily.Cell Mol Life Sci. 2008 Nov;65(22):3606-18. doi: 10.1007/s00018-008-8285-x. Cell Mol Life Sci. 2008. PMID: 18695941 Free PMC article. Review.
-
Crystal structures of the wild-type, P1A mutant, and inactivated malonate semialdehyde decarboxylase: a structural basis for the decarboxylase and hydratase activities.Biochemistry. 2005 Nov 15;44(45):14818-27. doi: 10.1021/bi051383m. Biochemistry. 2005. PMID: 16274229
-
Inactivation of malonate semialdehyde decarboxylase by 3-halopropiolates: evidence for hydratase activity.Biochemistry. 2005 Jul 5;44(26):9375-81. doi: 10.1021/bi050296r. Biochemistry. 2005. PMID: 15982004
-
The hydratase activity of malonate semialdehyde decarboxylase: mechanistic and evolutionary implications.J Am Chem Soc. 2004 Dec 8;126(48):15658-9. doi: 10.1021/ja044304n. J Am Chem Soc. 2004. PMID: 15571384
-
Evolution of enzymatic activity in the tautomerase superfamily: mechanistic and structural studies of the 1,3-dichloropropene catabolic enzymes.Bioorg Chem. 2004 Oct;32(5):376-92. doi: 10.1016/j.bioorg.2004.05.006. Bioorg Chem. 2004. PMID: 15381403 Review.
Cited by
-
The S. aureus 4-oxalocrotonate tautomerase SAR1376 enhances immune responses when fused to several antigens.Sci Rep. 2017 May 11;7(1):1745. doi: 10.1038/s41598-017-01421-z. Sci Rep. 2017. PMID: 28496136 Free PMC article.
-
Phylogenetic distribution of malonate semialdehyde decarboxylase (MSAD) genes among strains within the genus Mycobacterium: evidence of MSAD gene loss in the evolution of pathogenic mycobacteria.Front Microbiol. 2023 Oct 13;14:1275616. doi: 10.3389/fmicb.2023.1275616. eCollection 2023. Front Microbiol. 2023. PMID: 37901833 Free PMC article.
-
A global view of structure-function relationships in the tautomerase superfamily.J Biol Chem. 2018 Feb 16;293(7):2342-2357. doi: 10.1074/jbc.M117.815340. Epub 2017 Nov 28. J Biol Chem. 2018. PMID: 29184004 Free PMC article.
-
The structure of a tautomerase superfamily member linked to the type VI secretion system of Acinetobacter baumannii.Acta Crystallogr F Struct Biol Commun. 2023 Jan 1;79(Pt 1):8-16. doi: 10.1107/S2053230X22011414. Epub 2023 Jan 1. Acta Crystallogr F Struct Biol Commun. 2023. PMID: 36598351 Free PMC article.
-
Biochemical and Genetic Bases of Indole-3-Acetic Acid (Auxin Phytohormone) Degradation by the Plant-Growth-Promoting Rhizobacterium Paraburkholderia phytofirmans PsJN.Appl Environ Microbiol. 2016 Dec 15;83(1):e01991-16. doi: 10.1128/AEM.01991-16. Print 2017 Jan 1. Appl Environ Microbiol. 2016. PMID: 27795307 Free PMC article.
References
-
- Whitman CP, Aird BA, Gillespie WR, Stolowich NJ. J Am Chem Soc. 1991;113:3154–3162.
-
- Dagley S. Pathways for the utilization of organic growth substrates. In: Ornston LN, Sokatch JR, editors. The Bacteria: A Treatise on Structure and Function. Academic Press; New York: 1978. pp. 305–388.
-
- Murzin AG. Curr Opin Struct Biol. 1996;6:386–394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

