Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA
- PMID: 25219674
- PMCID: PMC4180118
- DOI: 10.1016/j.cell.2014.08.028
Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA
Abstract
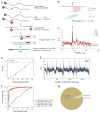
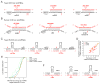
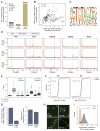
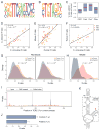
Pseudouridine is the most abundant RNA modification, yet except for a few well-studied cases, little is known about the modified positions and their function(s). Here, we develop Ψ-seq for transcriptome-wide quantitative mapping of pseudouridine. We validate Ψ-seq with spike-ins and de novo identification of previously reported positions and discover hundreds of unique sites in human and yeast mRNAs and snoRNAs. Perturbing pseudouridine synthases (PUS) uncovers which pseudouridine synthase modifies each site and their target sequence features. mRNA pseudouridinylation depends on both site-specific and snoRNA-guided pseudouridine synthases. Upon heat shock in yeast, Pus7p-mediated pseudouridylation is induced at >200 sites, and PUS7 deletion decreases the levels of otherwise pseudouridylated mRNA, suggesting a role in enhancing transcript stability. rRNA pseudouridine stoichiometries are conserved but reduced in cells from dyskeratosis congenita patients, where the PUS DKC1 is mutated. Our work identifies an enhanced, transcriptome-wide scope for pseudouridine and methods to dissect its underlying mechanisms and function.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

Comment in
-
Pseudouridine in a new era of RNA modifications.Cell Res. 2015 Feb;25(2):153-4. doi: 10.1038/cr.2014.143. Epub 2014 Nov 4. Cell Res. 2015. PMID: 25367125 Free PMC article.
-
Sequencing: Rereading familiar messages.Nat Methods. 2014 Nov;11(11):1095. doi: 10.1038/nmeth.3166. Nat Methods. 2014. PMID: 25551126 No abstract available.
References
-
- Bakin A, Lane BG, Ofengand J. Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry. 1994;33:13475–13483. - PubMed
-
- Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

