Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state
- PMID: 25219852
- PMCID: PMC4247276
- DOI: 10.1038/nature13722
Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state
Abstract
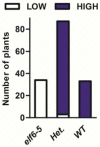
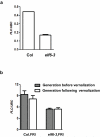
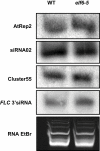
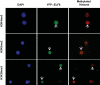
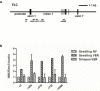
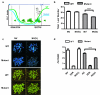
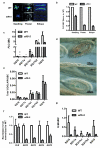
The reprogramming of epigenetic states in gametes and embryos is essential for correct development in plants and mammals. In plants, the germ line arises from somatic tissues of the flower, necessitating the erasure of chromatin modifications that have accumulated at specific loci during development or in response to external stimuli. If this process occurs inefficiently, it can lead to epigenetic states being inherited from one generation to the next. However, in most cases, accumulated epigenetic modifications are efficiently erased before the next generation. An important example of epigenetic reprogramming in plants is the resetting of the expression of the floral repressor locus FLC in Arabidopsis thaliana. FLC is epigenetically silenced by prolonged cold in a process called vernalization. However, the locus is reactivated before the completion of seed development, ensuring the requirement for vernalization in every generation. In contrast to our detailed understanding of the polycomb-mediated epigenetic silencing induced by vernalization, little is known about the mechanism involved in the reactivation of FLC. Here we show that a hypomorphic mutation in the jumonji-domain-containing protein ELF6 impaired the reactivation of FLC in reproductive tissues, leading to the inheritance of a partially vernalized state. ELF6 has H3K27me3 demethylase activity, and the mutation reduced this enzymatic activity in planta. Consistent with this, in the next generation of mutant plants, H3K27me3 levels at the FLC locus stayed higher, and FLC expression remained lower, than in the wild type. Our data reveal an ancient role for H3K27 demethylation in the reprogramming of epigenetic states in plant and mammalian embryos.
Figures


References
-
- Paszkowski J, Grossniklaus U. Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr. Opin. Plant Biol. 2011;14:195–203. - PubMed
-
- Becker C, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–9. - PubMed
-
- Mansour AA, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/00000581/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 233039/ERC_/European Research Council/International
- BBS/E/J/000CA369/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C517633/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/000CA376/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

